Introducción: La poliquistosis renal autosómica dominante (PQRAD) es la enfermedad hereditaria más frecuente con mayor amenaza para la vida. El análisis molecular de microsatélites polimórficos, localizados en torno a los genes responsables de la enfermedad (PKD1 y PKD2) se utiliza para confirmar el diagnóstico y dar consejo genético. Métodos: Se desarrolló un método de genotipado para cinco marcadores de PKD1 y cuatro de PKD2, basado en dos reacciones de PCR múltiple y análisis por electroforesis capilar. Un total de 110 individuos, pertenecientes a 14 familias afectas, fueron genotipados para confirmar la concordancia con el método de PCR simple usado previamente. Resultados: El rango de tamaño de los fragmentos amplificados fue desde 95 a 154 pb, y los perfiles completos de marcadores microsatélites se obtuvieron a partir de 1-5 ng de ADN. La especificidad de la PCR múltiple fue del 88,5% (IC 95% 75,9-95,2), y la sensibilidad del 87,9 (IC 95% 76,1-94,6). Conclusiones: Esta estrategia, junto con la detección automática de alelos, permite realizar un análisis genético fiable, sencillo, más rápido y de menor coste que el basado en amplificaciones individuales de los microsatélites.
Background: Autosomal Dominant Polycystic Kidney Disease (ADPKD) is the most common life-threatening hereditary disease. Molecular analysis with highly polymorphic short tandem repeats, located in the vicinity of the two genes responsible for the disease (PKD1 and PKD2), is used to confirm diagnosis and give genetic counseling to members of affected families. Methods: We have developed a new assay to genotype five PKD1 and four PKD2 markers, based on two multiplex PCR reactions, and capillary electrophoresis analysis. A total of 110 subjects, belonging to 14 affected families, were genotyped to confirm the concordance with the singleplex method used previously. Results: The amplicons ranged from 95 to 154 bp in length, and complete STR profiles were obtained from 1-5 ng DNA. The specificity of the multiplex PCR system was 88,5% (95%CI= 75,9-95,2), and the sensitivity, 87,9 (95%CI= 76,1-94,6). Conclusions: This is a useful strategy that, together with automated computer-based allele detection, allows reliable, simple, faster, and cheaper genetic analysis than the previous singleplex method.
INTRODUCTION
Autosomal dominant polycystic kidney disease (ADPKD) is the third most common cause of chronic kidney disease1 and the most life-threatening hereditary disease.2 Its prevalence varies between 1/400 and 1/1000 individuals, depending on the population.3 It is characterised by the formation of multiple cysts in the kidneys, and by cardiovascular and gastrointestinal disorders such as intracranial or aortic aneurysm, mitral valve prolapse, hernia of the abdominal wall, etc. 50% of these patients develop chronic kidney disease during their fifth decade.
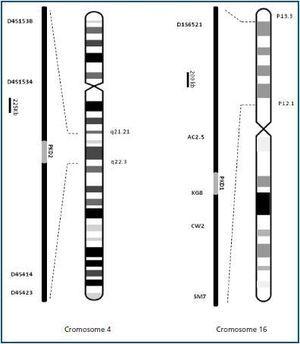
To date, two genes that cause the disease have been identified: PKD1, in chromosome region 16p13.3 (85% of all cases), and PKD2 in chromosome region 4q21 (15% of all cases). ADPKD is easily diagnosed with ultrasound when cysts are already present. However, genetic analysis can be used when ultrasound results are unclear or when a definitive diagnosis is needed for a patient younger than 30. An important use of molecular diagnosis is genetic counselling, which is the practice of providing the patient with information about his/her disease, the inheritance pattern and the risk of transmitting it to descendents (50%). It can also be used in families with ADPKD to confirm potential kidney donors. Clinical trials are currently being carried out to test the ability of some drugs to halt the progression of the disease.2 Patients who are diagnosed early may be candidates for treatment with these new agents in the future.
Molecular analysis can be done either through a direct search for mutations, or indirectly through gene binding analysis, using informative markers (microsatellites) located near the genes that interest us. These markers allow us to track the chromosome responsible for the disease through generations of the family being studied. Two microsatellite series selected in a preliminary study for PKD1 (D16S521, KG8, AC2.5, CW2, SM7) and PKD2 (D4S1538, D4S1534, D4S423, D4S414) were used for the gene binding analysis in families with ADPKD (figure 1). The suitability of these markers for our population was studied in 30 affected families.4 The markers were amplified individually and subsequently mixed for the analysis, which permitted unequivocal genetic identification of 97.7% in patients and 88.7% in healthy individuals. The sensitivity and specificity of the genetic analysis were 90.7% (CI 95%, 85.7-95.7) and 86.8% (CI 95%, 80.6-93.0) respectively.
In this study, we present a new analysis of PKD1 and PKD2 microsatellites based on multiplex-PCR and capillary electrophoresis.
MATERIAL AND METHOD
A total of 110 individuals, 52 affected by the disease (57.7% male) and 58 not affected (39.7% male) were analysed using the new multiplex-PCR system. These individuals belonged to 14 families with ADPKD who were randomly selected from those that were included in the preliminary analysis.4 The study was approved by the Ethics Committee at Dr Negrín University Hospital of Gran Canaria. Furthermore, we obtained informed consent from all participants in the study before they were included in it.
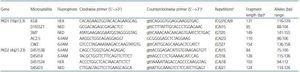
A new series of primers was designed for developing the multiplex-PCR, which ensured equivalent amplification efficiency for all DNA fragments, as described previously.5 The length of the amplified fragments varied between 75 and 158 base pairs (table 1). The objective was to obtain primers with a GC content of about 35-60% and a theoretical melting temperature of 60 ± 2∫ C in a salt concentration (K+, Na+, Tris+ or NH4+) of 180mM. As a primer-binding site, complex sequences, homopolymers with more than five bases and potentially stable secondary structures (below the threshold of -13kcal mol-1 of free energy) in order to guarantee the sensitivity and specificity of the multiplex amplification.6 We used the program available at Integrated DNA Technologies (http://www.idtdna.com/Scitools/Applications/mFold/) to verify that none of the candidate strains presented secondary structures or hairpins. The strains were synthesised and purified using HPLC by Bioron GmbH (Ludwigshafen
Germany) and Applied Biosystems (Foster City, U.S.A). To differentiate amplified products of similar sizes, we used a multi-colour fluorescent and marked the clockwise primer for each pair with one of the following fluorophores: 6- FAM, HEX or NED. Each pair of primers was tested in an individual PCR with five DNA samples, under identical amplification conditions, before carrying out the multiplex PCR reaction. The quantity of each primer included in the final mixture was established empirically, in accordance with results from consecutive tests. Furthermore, different series dilutions were tested for MgCl2, between 1.5 and 5mM, in order to obtain the maximum amplification efficiency.
Owing to a slight overlap between the size ranges for CW2 and D4S414 in some samples, we decided to analyse PKD1 and PKD2 markers separately. Both reactions contained 1-10ng of DNA, 3mM MgCl2, 200μM of each dNTP, 0.05-0.1μM of each primer and 0.5U AmpliTaq Gold polymerase (Applied Biosystems) in a final reaction volume of 12.5μl. The amplification protocol was carried out in a GeneAmp PCR System 9700 Thermal Cycler (Applied Biosystems), and consisted of denaturalisation at 95∫ C during 5 minutes, followed by 26 cycles of 95∫ C during 30 seconds, 65∫ C during 30 seconds and 72∫ C during 45 seconds, with a final extension step of 20 minutes at 72∫ C. A product of the desired size was obtained for all loci. In order to analyse the robustness of multiplex-PCR, duplicate analyses were run for each sample.
Detection of PCR fragments using capillary electrophoresis was carried out in an ABI TM 310 sequencer (Applied Biosystems), using an uncovered capillary 47cm long and POP4 polymer (Applied Biosystems). For the electrophoresis, 1μl of each PCR product was mixed with 20μl de-ionised formamide (Applied Biosystems) and 0.2μl of GeneScan-ROX molecular weight marker (Applied Biosystems). Samples were injected with 15kV during 5 seconds, and separated with the same voltage and a temperature of 60∫ C during 24 minutes.We used the software GeneScan Analysis 3.1.2 (Applied Biosystems) to estimate fragment size.
RESULTS
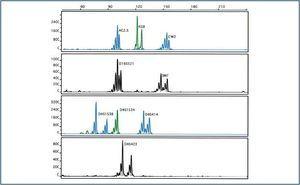
Initially, four markers (D4S423, CW2, D16S521 and SM7) gave off a weak signal, which suggested poor amplification under the same conditions and with the same quantities of primer. Reactions improved when primer quantities increased, thus reaching the desired balance between signals. The best results were obtained with a concentration of 3mM MgCl2 for all markers (figure 2). Slightly higher concentrations inhibited amplification.
Identification of individual haplotypes showed 100% concordance with the single-PCR results gathered prior to the analysis. Markers were completely informative for all patients and in 91.4% of the healthy individuals. Multiplex-PCR demonstrated a specificity of 88.5% (CI 95%, 75.9-95.2) and a sensitivity of 87.9 (CI 95%, 76.1-94.6).
DISCUSSION
A large number of markers for analysing the PKD1 and PKD2 genes have been described. In our preliminary study,4 we used a simple PCR system to amplify two series of microsatellite PKD1 and PKD2 markers. To develop this technique, it was necessary to run nine different PCR reactions for each individual, mix the amplification products, and lastly, prepare the sample for capillary electrophoresis. Until now, the simplest multiplex-PCR system published for ADPKD analysis was the one by Vouk et al,7 which consists of three amplification reactions, combining PKD1 and PKD2 markers.
An important part of our work consisted of exploring various aspects of multiplex-PCR methods. Primers were designed paying special attention to avoid self-alignment and dimer formation, obtaining maximum efficiency and balance between the quantities of amplification products. The primers were subjected to HPLC in order to obtain pure, homogeneous molecules, without a surplus of fluorophores that might interfere with allele detection. To avoid non-specific amplification products, we used the PCR programme “Hot Start” (in which the polymerase is activated at high temperatures and the PCR products accumulate slowly).
We have developed a new multiplex-PCR system for genotyping nine microsatellite ADPKD markers in only two reactions. An additional advantage of this system is that it permits independent analysis of PKD1 and PKD2. Given that 85-90% of ADPKD cases are linked to PKD1, analysing the markers for this gene will be sufficient in order to determine the family’s genetic characteristics in many cases. Genetic analysis using the multiplex-PCR system affords greater ease in both genetic counselling and early diagnosis, which is increasingly important for this disease. To conclude, this strategy, in conjunction with automatic allele detection, allows us to carry out a reliable, simple genetic analysis that is faster and less expensive than the one based on individual amplifications of microsatellites, which encourages including family studies in the clinical routine.
Figure 1.
Table 1. Microsatellites and primers
Figure 2.