Extracellular vesicles (EV) reflect the pathophysiological state of their cells of origin and are a reservoir of renal information accessible in urine. When biopsy is not an option, EV present themselves as sentinels of function and damage, providing a non-invasive approach. However, the analysis of EV in urine requires prior isolation, which slows down and hinders transition into clinical practice. The aim of this study is to show the applicability of the “single particle interferometric reflectance imaging sensor” (SP-IRIS) technology through the ExoView® platform for the direct analysis of urine EV and proteins involved in renal function.
Materials and methodsThe ExoView® technology enables the quantification and phenotyping of EV present in urine and the quantification of their membrane and internal proteins. We have applied this technology to the quantification of urinary EV and their proteins with renal tubular expression, amnionless (AMN) and secreted frizzled-related protein 1 (SFRP1), using only 5 μl of urine. Tubular expression was confirmed by immunohistochemistry.
ResultsThe mean size of the EV analysed was 59 ± 16 nm for those captured by tetraspanin CD63, 61 ± 16 nm for those captured by tetraspanin CD81, and 59 ± 10 for tetraspanin CD9, with CD63 being the majority EV subpopulation in urine (48.92%). The distribution of AMN and SFRP1 in the three capture tetraspanins turned out to be similar for both proteins, being expressed mainly in CD63 (48.23% for AMN and 52.1% for SFRP1).
ConclusionsThis work demonstrates the applicability and advantages of the ExoView® technique for the direct analysis of urine EV and their protein content in relation to the renal tubule. The use of minimum volumes, 5 μl, and the total analysis time not exceeding three hours facilitate the transition of EV into daily clinical practice as sources of diagnostic information.
Las vesículas extracelulares (VEs) reflejan el estado fisiopatológico de sus células de origen y constituyen un reservorio de información renal accesible en orina. Cuando la biopsia no es una opción, las VEs se presentan como un centinela de funcionalidad y daño, constituyendo una aproximación no invasiva. Sin embargo, el análisis de las VEs de la orina requiere de un aislamiento previo que ralentiza y dificulta su traslación a la práctica clínica. El objetivo de este trabajo es mostrar la aplicabilidad de la tecnología “sensor de imágenes de reflectancia interferométrica de una sola partícula” (SP-IRIS) mediante la plataforma ExoView® para el análisis directo de las VEs de la orina y de proteínas implicadas en funcionalidad renal.
Materiales y métodosLa tecnología ExoView® permite la cuantificación y fenotipado de las VEs presentes en orina y la cuantificación de sus proteínas, de membrana e internas. En este trabajo se aplica esta tecnología a la cuantificación de las VEs de la orina y de sus proteínas con expresión renal tubular, amnionless (AMN) y secreted frizzled-related protein 1 (SFRP1), empleando únicamente 5 μL de orina. La expresión tubular se confirmó por inmunohistoquímica.
Resultados. El tamaño medio de las VEs analizadas fue de 59 ± 16 nm para las capturadas por la tetraspanina CD63, 61 ± 16 nm para las capturadas por la tetraspanina CD81 y 59 ± 10 para la tetraspanina CD9, siendo CD63 la subpoblación de VEs mayoritaria en orina (48,92%). La distribución de AMN y SFRP1 en las 3 tetraspaninas de captura resultó ser similar para ambas proteínas, expresándose y mayoritariamente en CD63 (48,23% para AMN y 52,1% para SFRP1).
Conclusiones. Este trabajo evidencia la aplicabilidad y ventajas de la técnica ExoView® para el análisis directo de las VEs de la orina y su cargo proteico en relación con el túbulo renal. El empleo de volúmenes mínimos, 5 μL, y el tiempo total de análisis no superior a 3 horas facilita la traslación a la práctica clínica diaria de las VEs como fuentes de información diagnóstica.
Extracellular vesicles (EVs) reflect the content and pathophysiological state of the cells where they originate and therefore constitute a slot of study of great potential in clinical practice. They contain nucleic acids, proteins, metabolites and lipids that can be transfered to other cells from the cell of origin, thus mediating cellular communication.1 Initially considered as a cell waste product, however the interest in EVs has been increasing in recent years. Particularly if tissue (biopsy) is inaccessible in routine clinical procedures, EVs become a lookout of function and damage, being also a very valuable reservoir of biomarkers that are obtained non-invasively.2–4 Their interest also lies in the fact that they are not only intercellular communicators of variable proximity, but by migrating through the blood they are also mediators between organs and systems of the organism. Furthermore, given their capacity to be internalized by target cells, they have been more recently proposed as vehicles for therapeutic action to activate or inhibit mechanisms of interest once their function has been completed.5,6
Within the nephrological context, the urine EVs reflect the pathophysiologic state of the kidney, and they stablish intercommunications within the glomerulus, the tubule, and both functional structures with the tubulointerstitial space.7,8 In 2004, the presence of renal system proteins in urine EVs was first evidenced by mass spectrometry.9 Subsequent studies using omics approaches have confirmed this finding and the correlation between the protein levels detected in EVs and in the kidney.10 Several studies focused on acute renal failure, polycystic kidney disease, glomerular disease and tubulopathies have demonstrated their potential as a non-invasive source of biomarkers, showing how their excretion and/or content are conditioned by nephron mass.11 Therefore, it is clear that the analysis of urine EVs allows access to molecular alterations taking place in the kidney, without the need to perform a biopsy.12 Such alterations could even reflect subclinical organic damage not detectable in routine follow-up or diagnostic analyses. Apart from its biological interest, at a methodological level, the study of urine EVs has a great advantage over direct urine analysis, since patients with high level of proteinuria will have proteins of interest masked because they are at low levels of abundance with respect to the predominant proteins (such as albumin). However, the proteins present in the EVs (cytosolic and membrane) purified appropriately, represent their cells of origin and will see this interference eliminated, thus increasing the possibilities of quantifying proteins at lower levels of abundance and have access to new biomarkers.13
The main drawback to implement its diagnostic application in clinical practice in the near future is that its analysis implies a previous isolation step, usually performed by ultracentrifugation or size exclusion chromatography, which implies potential sample alterations and a long duration analysis, thus making direct translation impossible. Recently, the SP-IRIS technology implemented in the ExoView® equipment (Unchained labs, USA) allows the quantification and direct phenotyping of EVs present in any diagnostic fluid, as well as the quantification of protein biomarkers located both in its membrane and inside.14,15 Its main advantage lies, therefore, in performing the analysis of EVs directly in the biological fluid of interest without prior isolation. Moreover, it will do so using minimal sample volumes and in a short period of time.
In this article we show the potential of ExoView® technology in the direct, targeted and simultaneous analysis of 2 renal proteins in urine EVs.
MethodsTwo proteins of interest with tubular expression were selected based on previous knowledge of the laboratory and the Human Protein Atlas database: amnionless (AMN), located in the membrane of the EVs, and secreted frizzled-related protein 1 (SFRP1), part of the internal content. Their tubular location was analysed by immunohistochemistry in human renal tissue obtained from the Fundación Jiménez Díaz Biobank, using 5 μm sections and the anti-AMN (HPA000817, Merck Millipore) and anti-SFRP1 (PA5-95634, Invitrogen™) antibodies.
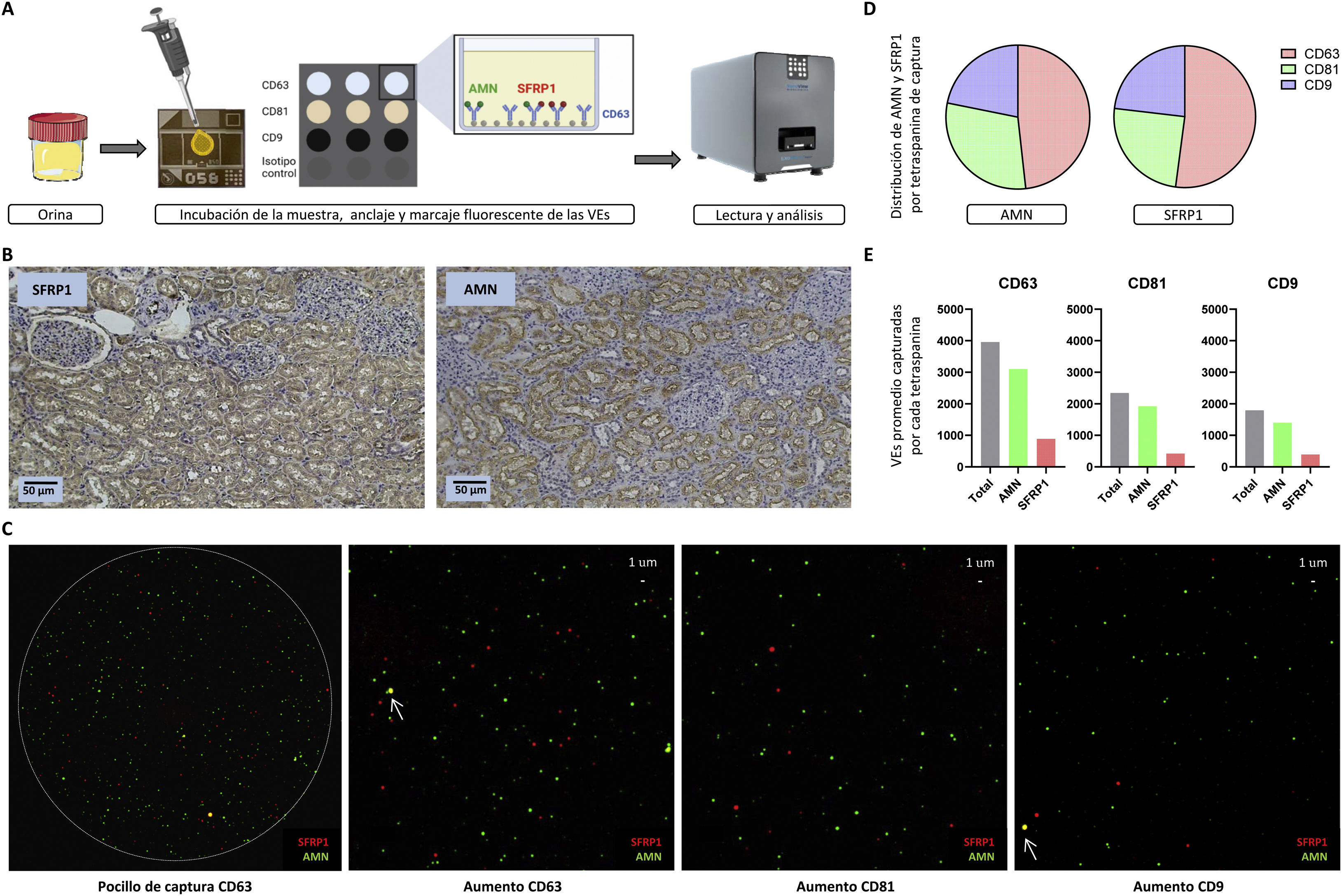
Urine EVs were analysed by ExoView® technology using a standard commercial tetraspanin CD63, CD81 and CD9 capture kit (EV-TETRA-C, Unchained labs) through antibodies for human samples (anti-AMN; anti-SFRP1). The analysis of these EVs in urine was performed following the manufacturer's instructions. First, a pre-scan of the capture medium (chip) was performed as a quality control and blank in the data analysis. Then, the urine concentration to be used in the analysis was optimized. Without pre-isolating the EVs, 50 μl of diluted urine was directly loaded into incubation dilution and incubated on the chip in the dark at room temperature overnight. Alternatively, this incubation can be carried out in 1−2 h without afecting the results in case it is necessary to significantly reduce the time of analysis. For the detection of intracellular proteins such as SFRP1, a permeabilization step was included that would not be necessary for a membrane protein analysis of EVs. EVs present in urine anchored to the chip through their tetraspanins were incubated in shaking and darkness for 1 h with anti-AMN (CF 594A) and anti-SFRP1 (Dylight 755) labeled antibodies using Alexa Fluor Conjugation Kits Fast (Abcam). Finally, the chip was read in the ExoView® Analyzer (Unchained labs) to detect, localize and quantify the proteins of interest in each of the EVs. Each chip has 3 capture wells per tetraspanin. In this way, it can be obtained both total values (sum of the 3 wells of each tetraspanin) and average values, in terms of EVs captured and proteins analysed. Fig. 1A summarizes the workflow.
(A) ExoView® workflow. Urine, without previous isolation of EVs, is added on the central part of the chip, which contains the wells where the EVs are captured by the different tetraspanins (CD63, CD81 and CD9). These EVs are fluorescently labeled in green if they express AMN or in red if they express SFRP1. The chips are read and analysed in the ExoView® R200. (B) Analysis of AMN and SFRP1 in human renal tissue by immunohistochemistry. Both glomeruli and tubules are visible (×10 magnification), with only the latter being stained, as they are the functional structures related to our proteins of interest. (C) Visualization of the capture of the proteins of interest (AMN, SFRP1). An image of one of the wells of the CD63 capture tetraspanin is shown, in which the specific and delimited binding of EVs can be appreciated. On the right, a magnification of a well for each capture tetraspanin (CD63, CD81 and CD9, respectively) is included, with each coloured dot being a vesicle expressing AMN (membrane protein, in green) or SFRP1 (intracellular protein, in red). In addition, the co-localization of both proteins (yellow) in the same EV is indicated by arrows. (D) Distribution (%) of AMN and SFRP1 proteins by capture tetraspanin (CD63 in red, CD81 in green and CD9 in blue). (E) Quantification of captured EVs containing either of the two studied proteins (AMN in green and SFRP1 in red), for each of the tetraspanins (CD63, CD81 and CD9). Average values are shown for the 3 wells for each capture tetraspanin.
Immunohistochemical analysis shows the tubular localization of the 2 proteins analysed, SFRP1 and AMN (Fig. 1B). The appropriate dilution for analysis of the urine samples by ExoView® was set at 1:10, so that only 5 μl of urine were necessary for analysis. A total of 24,297 EVs were captured, of which 48.92% were captured by CD63, 28.93% by CD81 and 22.15% by CD9, thus CD63 being the majority EV subpopulation in urine. This is in line with the participation of this subpopulation of EVs in tubular functionality, since the expression of this tetraspanin has been related to the functionality of the proximal tubule.16 The mean size of the EVs analysed was 59 ± 16 nm for those captured by CD63, 61 ± 16 nm for those captured by CD81 and 59 ± 10 nm for CD9; corresponding in all 3 cases to small-sized EVs.
Fig. 1C shows a visual representation of the EVs analysed highlighting the detection of AMN (green) and SFRP1 (red) by the different capture tetraspanins (CD63, CD81 and CD9). Considering only EVs expressing each of the proteins of interest analysed, the distribution of AMN and SFRP1 in the different tetraspanin subpopulations was calculated. A 48.23% of AMN is detected in the total of EVs captured by CD63, 29.92% in EVs captured by CD81 and 21.85% in those captured by CD9. The distribution of SFRP1 is similar: 52.10%, 24.77% and 23.13%, respectively (Fig. 1D). To quantify the proteins of interest, we assessed the detection of each protein in EVs captured by each tetraspanin (average values). We found that of the 3,962 average EVs captured by CD63, 78.29% were positive for AMN and 22.46% for SFRP1. In the case of CD81, of the 2,343 average EVs captured, 82.07% were positive for AMN and 18.05% for SFRP1. Finally, on CD9 we found an average of 1,793 EVs captured, with AMN expressing 78.30% and SFRP1 22.03% (Fig. 1E). The total percentages do not necessarily equal 100%, as some of the EVs may contain both proteins. These analysis show how for these 2 proteins there is a higher concentration of EVs containing AMN than SFRP1 and yet the distribution of the proteins in the different subpopulations of EVs is similar.
It is believed that there are separate populations of EVs specialized in different pathways and biological functions.17 Until now this has been difficult to demonstrate as it requires studies that uses technology able to assess EVs individually. In the study of a possible biological function of specific subpopulations of tetraspanins, ExoView® also makes possible to analyse the co-localization (co-expression) of the proteins of interest, which, if mainly present in a subpopulation, could be related to their participation in a specific functional structure. Analysing the co-localization of AMN and SFRP1 in EVs captured by the different tetraspanin subpopulations, we found low co-expression of both proteins in the same vesicle; 0.73% in the case of vesicles captured by CD63, 0.13% for EVs captured by CD81 and 0.33% on CD9.
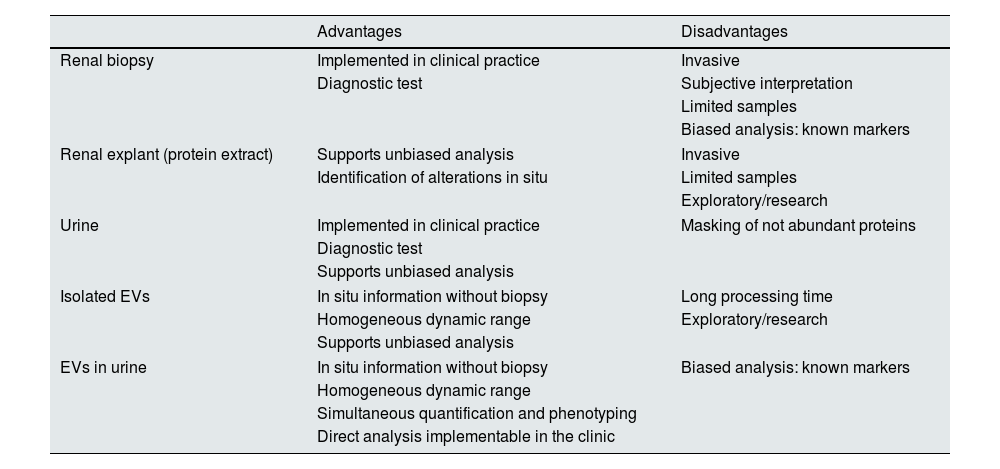
In this study, two urine EV proteins expressed in the tubule were investigated, demonstrating the advantages of ExoView® analysis for the characterization and direct quantification of EVs, which can be applied to other proteins and in other biological fluids of clinical interest. Table 1 shows a comparison of the different types of biological samples that can be analyzed for the study of renal diseases with their advantages and disadvantages in terms of the information they provide, their availability and their possibility of implementation in health systems. In ExoView®, the use of volumes as small as 5 μl and total analysis time of no more than 3 h facilitates the translation to daily clinical practice of EVs as sources of diagnostic information. In the context of renal diseases, EVs enable the monitoring of markers of renal function, therapeutic efficacy or estimation of cardio-renal risk.18,19 Along these lines, the technique should be validated in large, multicentre cohorts in order to confirm its potential in clinical diagnosis and its application to routine clinical analyses.
Types of biological samples that can be evaluated for the study of renal disease.
| Advantages | Disadvantages | |
|---|---|---|
| Renal biopsy | Implemented in clinical practice | Invasive |
| Diagnostic test | Subjective interpretation | |
| Limited samples | ||
| Biased analysis: known markers | ||
| Renal explant (protein extract) | Supports unbiased analysis | Invasive |
| Identification of alterations in situ | Limited samples | |
| Exploratory/research | ||
| Urine | Implemented in clinical practice | Masking of not abundant proteins |
| Diagnostic test | ||
| Supports unbiased analysis | ||
| Isolated EVs | In situ information without biopsy | Long processing time |
| Homogeneous dynamic range | Exploratory/research | |
| Supports unbiased analysis | ||
| EVs in urine | In situ information without biopsy | Biased analysis: known markers |
| Homogeneous dynamic range | ||
| Simultaneous quantification and phenotyping | ||
| Direct analysis implementable in the clinic | ||
EVs: extracellular vesicles.
The research activity of the authors has been funded by Insituto de Salud Carlos III through the projects (PI20/01103, CP22/00100, IF08/3667-1, RD16/0009, RD21/0005/0001 (Co-funded by European Regional Development Fund/European Social Fund “A way to make Europe”/“Investing in your future”) and the Community of Madrid (PEJ-2020-AI/BMD-17899, PEJD-2019-PRE/BMD-16992, 2018-T2/BMD-11561). This study has been funded primarily by the SENEFRO Foundation and partially by the Mutua Madrileña Foundation and the Spanish Proteomics Society.
Conflict of interestThe authors have no conflicts of interest.