INTRODUCTION
The publication of a series of articles in the January issue of the journal JAMA titled ¿How to use an article about genetic association¿1-3 serves as an excellent starting point to try a two-fold purpose: to present, similarly to what has been published,a practical guide with the necessary requirements to face an article on genetic association, and, on the other hand, to show the necessary tools to perform such a study. To the present purpose, we reduce the scope of action to those works on population based genetic association that are carried out by recruiting cases and controls considering that we will be evaluating candidate genes. Methods and interpretation of results from family-based genetic studies are different and are not within the scope of this revision. We insist in that our intention is only to point out a series of practical application guidelines and not to attempt any approach to genetic epidemiology. We will therefore draw some considerations over the clinical relevance of these studies and analyse the current situation and its survival against genome-wide association studies.
ARE THE PATIENTS APPROPRIATELY SELECTED? PHENOTYPIC CHARACTERISATION
The adequate characterisation of a phenotype associated with certain disease is to be done in compliance with those clinical criteria over which there is a clearly established medical-scientific agreement. Most scientific societies set these criteria and, of course, their corresponding updates, as knowledge about disease progression and evolution is enhanced. In real terms, however, it is not always possible to establish the right phenotype even when the guideline criteria are rigorously followed.
IS SAMPLE SIZE THE RIGHT ONE?
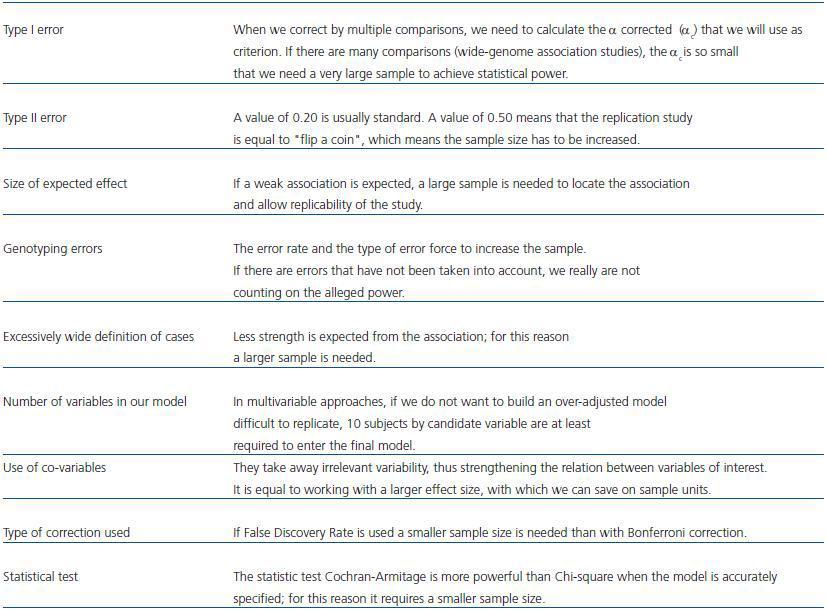
Assessing sample size (table 1) in a case-control study that includes genetic information is an object of constant study and remodeling.4 A usual approach to assessing the sample size in a genetic association study does not differ from one in a usual clinical exposed/not exposed-type study, and it is based on previously establishing the magnitude of the difference to detect. In our particular case, it would mean to establish a priori the difference between the allelic or genotypic frequency in our populations. In addition, we have to know the frequency of alleles (all those to be considered) in the control population, the value of type I error, i.e. the error of rejecting the null hypothesis when the null hypothesis is true, and type II error, that is accepting the null hypothesis when the null hypothesis is false. We usually play safe as not to make type I error in 95% (probability, α = 0.05) and 5 to 20% type II; although, it is usually set at 20% (probability, β = 0.2). Thus a statistical power (1 - β) of 80% is ensured. Other aspects should be considered too, such as predictable error rate and type of errors to be expected in our genotyping procedure, which should be compensated by a larger sample size so as not to reduce the statistical power. Some ¿on-line¿ tools that help us in these calculations are, among others: http://linkage.rockefeller.edu/pawe/ y http://hydra.usc.edu/GxE/.
DISCUSSION OVER THE CASES
Ideally, incident cases should be recruited. Knowing in advance the allelic frequency of the variants to be studied and before proceeding to recruiting phase, and being aware of the difficulty inherent in a given phenotype, the number of subjects (cases) to be studied could be increased through various alternatives: on the one hand, by increasing the number while being less demanding when defining the phenotype, as for example in «renal disease», thus increasing heterogeneity and reducing certainty to allele causality under evaluation; on the other hand, phenotype selection criteria could be more rigorous, thus increasing homogeneity but necessarily reducing the number of cases recruited. Alternatively, phase duration for recruitment could be increased. Choosing one alternative depends on various factors, mainly on the allelic frequency of the variation(s) in the candidate gene(s) to be studied. If an open criterion is chosen to select the cases, it is to be expected that the candidate genes are present in a lower sub-group of our cases to study; therefore, due to the smaller effect size expected we will at least lose part of the statistical power that we intended to attain when defining our phenotype more broadly. Hence, when it comes to defining the cases we have to consider whether incrementing the cases available compensates for the loss in statistical power derived from a smaller difference in the frequencies expected. If we postpone defining the phenotype until the moment when we have performed the analysis of our results, we have alternatives. Some authors, such as Chen and Lee, have created a simple quantitative method that allows us to clarify and systematize, depending on the allelic frequencies and the existence of at least two possible types of «cases», when we can increase or not the size of our sample by linking both case types.5 Nevertheless, it is most scientifically appropriate to act in advance of these questions at the design phase of the study and not when we proceed to perform the statistical analysis, since we might over-adjust the data thus obtaining spurious results that are difficult to replicate.
DISCUSSION OVER THE CONTROLS
Arya Sharma and Xavier Jeunemaitre,6 authors of renown in this area, point at a common difficulty and error in the selection and control population recruitment. The difficulty stems from the very nature of the control population. While for most of the hospital researchers the inclusion of patients has not been a problem, control recruiting has, as it requires a population basis with specific resources. Given that a control is potentially a case, a usual bias is to include controls from, for example, blood banks or healthy workers in the area. Selecting these candidates defined as «hyper normal» would theoretically result in a major difference in allelic frequencies between the affected and the control population, but the advantage to this is indeed minimal and it might hide other phenotypes of positive selection toward survival.7-9 Likewise, selecting controls that are not diagnosed cases entails a reduction in the statistic power of our study.10
ON STATISTICAL ANALYSIS AND STATISTICAL ASSOCIATION
We can set a phenotype selection that has to be studied characterising the phenotype as a dichotomous trait: diabetics against non-diabetics, hypertensive against normotensive, or by trying to further delimit variability by reducing environmental uncertainty and gaining in phenotype influence through the evaluation of so-called intermediate phenotypes, i.e. by evaluating one or many measurable characters that linked by plausible biological pathways our candidate gene(s) to the disease. The most conventional statistical approach to either one alternative implies carrying out multivariate models.
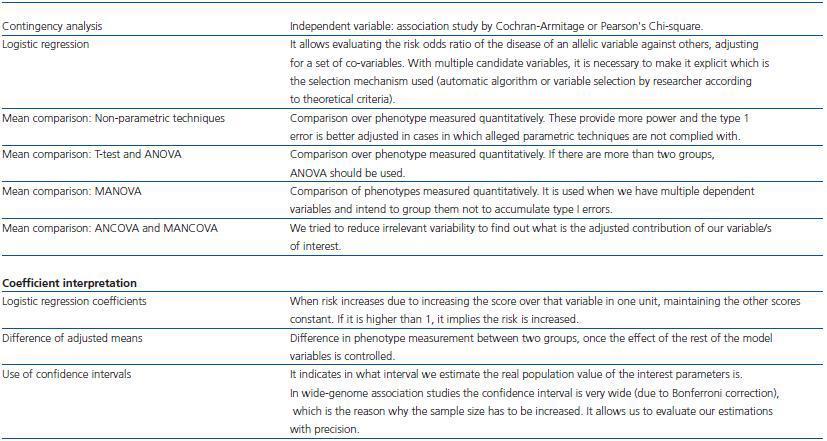
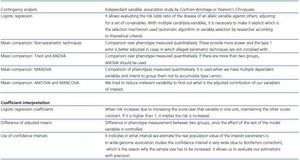
In studies with multiple comparisons we have to use corrections to avoid increasing type I error (table 2). Bonferroni procedure consists in dividing alpha by the number of comparisons to estimate: if there are approximately 106 variants in the genome, the threshold value of p corrected for all the comparisons would be p = 0.05/106 = 5 X 10-8. If we intend to detect moderate differences this forces us to have a large sample size. For this reason, less conservative correction formulas are used, such as the False Discovery Rate. However, when making numerous comparisons even using this procedure requires increasing the sample size too much.
The statistical analysis used may give us a major statistical power (table 3). Thus, if we know the genetic model (additive-recessive-dominant), we should use the Cochran-Armitage test. However, in general we do not know the genetic model of our candidate genes, and although it gives us more power, it is also true that it turns out to be less robust than the traditional Pearson¿s chi-square test, which is the reason why in case of non-compliance with the cases we set up over the genetic model the results would become invalidated.
ANALYSIS OF POPULATION MIX
The need, in medicine, to evaluate genetic heterogeneity in the population under study derives from the current evolution theory. The studies show that although there is variation of frequency between alleles associated with the disease between dissimilar populations, this is indeed quite small.7 It might be the case that the association depended wholly on the exposure to certain environmental determinant whose frequency varied according to geographic localization and that frequency of these alleles by selection varied accordingly.7 When population mixes are made that differ in allelic frequencies because of genetic or environmental reasons the association may turn out to be spurious. Therefore the need for selection, genotyping, and neutral markers analysis (null alleles, unlinked SNPs, insertions/deletions) through two different strategies termed ¿genomic control¿ and ¿structured association¿: http://pritch.bsd.uchicago.edu/structure.html and http://wpicr.wpic.pitt.edu/WPICCompGen/genomic_control/genomic_control.htm
SELECTION OF CANDIDATE GENES
Since in our approach we have decided to evaluate gene polymorphisms in candidates genes, we are interested, firstly, in determining both the number and what genes we analyse. The candidate genes under analysis are traditionally selected based on of the knowledge about the following: gene product activity in the disease under study, function of the protein coded by such gene, information on studies in animal models, knowledge presupposed from the phenotype associated with monogenic forms of the disease, knowledge derived from gene linkage studies, and data available a priori, as well as knowledge obtained from meta-analysis.8 If, in addition, the variants to be analysed are located in regions of interest in the genes, this translates, for example, in a change of amino acid (non-synonymous variants), or they affect the stability of the messenger processing; or if the variant is located in regulating regions of the gene, then that variant would probably be more useful.11 Another strategy in the selection of candidate genes is the selection of tag variants, i.e. variants on which there is prior information about, information obtained by linkage studies and which at the same time can be linked to susceptibility alleles.12
In such an active area as it is bioinformatics, it is surprising that there is such a small number of tools aiding a key task as it is the selection of candidate genes.13 There are, however, a number of on-line programmes: http://omicspace.riken.jp/PosMed/,http://www.genesniffer.org/index/index_frameset.htm and http://www.genetics.med.ed.ac.uk/suspects/. These programmes are part of the information from platforms of high performance genetic analysis together with the information from expression studies allowing the appearance of the term convergence to region selection and candidate genes.
QUANTIFIABLE PHENOTYPE AND EXPERIMENTAL DEMONSTRATION OF QUANTIFIABLE PHENOTYPE
Identifying and measuring the number of biologic parameters involved directly with the gene and its product or with the biologic pathway in which it is involved significantly increases the study information capacity. It is more rigorous and, in addition, it is experimentally demonstrated that the variable itself associates with other variables in key regions of the gene or that it functionally affects either the gene or the protein.6
DOES IT MAKE SENSE TO STUDY AND ANALYSE A RELATIVELY SMALL NUMBER OF POLYMORPHISMS?
Analysing just one polymorphism can lead to spurious associations, among other things, because the variant may be at linkage disequilibrium with other variants, thus forming a characteristic haplotype. Sharma, et al.6 consider that for a candidate gene, selection, genotyping, and frequency analysis of, at least, three common polymorphisms allows identifying variants at linkage disequilibrium and identifying synergies. In a population-based case-control study where proximal SNPs have been genotyped and given that by definition the phase is unknown, haplotypes may be inferred by using genetic software applications: (GDA: http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php and Arlequín: http://lgb.unige.ch/arlequin/), but it should not be determined in its totality, neither should it be associated with a measurable phenotype in a give subject (except those genotyped as homozygous for all the SNPs evaluated in a locus). Nowadays, information on haplotypes begins to be available in the Internet and it is therefore interesting to choose and validate the predictive or therapeutic usefulness of functional variants.
Positive associations found in a given population using one or a few variants do not usually replicate in other populations. Lack of replication is a fundamental argument against association studies amongst the most critical authors. Apart from the problems derived from genotyping errors, population mix, election of candidate genes, inadequate recruiting and characterization of cases and/or controls, and differences in environmental exposure, there is lack of statistical power with absence of association (table 4) in most of the studies. The solution is not simple, and neither is it recruiting a larger number of cases and controls.
A retrospective case-control genetic association study should therefore comply with a series of requirements succinctly presented below.
LARGE SCALE GENOTYPING AND GENOME-WIDE OR GENOME-WHOLE ASSOCIATION STUDIES
The International HapMap Project (http://www.hapmap.org/index.html.en) is defined as a joint effort from many countries to identify and catalogue genetic similarities and differences in human beings at this level.14 As mentioned above, the methodology and interpretation of the results obtained differs from analysis of candidate genes in unrelated individuals. High-performance genetic analysis platforms have changed the view of the association studies by making it possible to genotype multiple polymorphisms, although necessarily enhancing the size of the sample. Large-scale merging of genotyping technologies with the available information at the International HapMap Project has provided the possibility to perform genetic association studies using information termed as whole-genome association or wide-genome association (WGA). In these studies HapMap provides the information of the so-called ¿tag SNPs¿, defined as the minimum set of SNPs needed to detect a haplotype. The project has gone through phases. Trios were recruited in the first phase (mother, father, and children) from whom SNPs located at the distance of 5kb and with a frequency > 5% were identified. A haplotype-like structure was then characterised to define the ¿tags¿: In a second phase, identifying ¿tags¿ associated with a certain disease allows inferring the haplotype structure, thus reducing the need to genotype all the variants and making it possible to locate the next candidate genes. These types of studies, although increasingly affordable, today require significant human and financial resources, but they are also an important step toward characterising clinically relevant variables. The technology associated with genotyping is not however free from problems.15 Even when an endeavour of such magnitude is carried out a few variants statistically associated with the disease survive the process of replication.16 An added problem is the need of correction when multiple statistical comparisons are performed9,10.
CLINICAL USEFULNESS
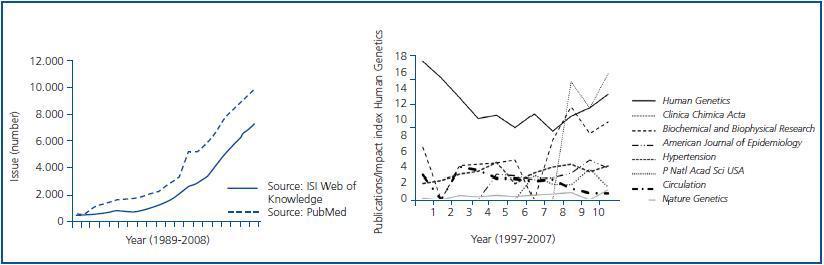
The initial enthusiasm caused by the genetic association studies was based on how easily these allowed going a step further over the conventional epidemiological approach in the knowledge of disease causality and/or risk factors associated with the disease (figure 1). For one or many candidate genes, most health centres were able, for example, to carry out PCR amplifications and enzymatic digestion of a series of polymorphisms in genes of interest once their cases and controls had been recruited. However, this single information remained biased. This relative ease allowed an exponential growth in the number of publications and, concomitantly, the appearance of a sector critical to its usefulness. Some authors point out, additionally, that as the action or effect of certain variant within a gene must be interpreted within the context of a complex net including, apart from interactions with other variants and the environment, the complexity itself of the biological pathway where the gene is embedded, the validity of an association strategy should be set out in the beginning.7 In many studies carried out initially the main critical point was the lack of reproducibility in other series and populations. However, it is worth noting that the common denominator that in many cases justifies the lack of reproducibility does not depend so much on the population under analysis but, as already remarked, on the lack of statistical power, which becomes the principal drawback. Since the emerging technology allows increasing exponentially the number of variations to analyse, good population recruitment becomes both the main requirement and a major problem. In Spain, the law of biomedical investigation 14/2007 of 3 July, regulates the type of genetic studies that can be performed, the structure of informed consent necessary, anonymisation process of samples and storage, utilisation, and transfer. Consequently, the following points are succinctly related to what has been mentioned above: inadequate characterisation of the population to be studied, lack of an adequate evaluation of the population, inadequate recruitment of cases and/or controls, insufficient size of sample, and lack of replication from analysed associations. Certain scepticism arises that can, however, be counteracted with characterisation of adequate phenotypes, analysis of intermediate phenotypes, evaluation of measurable phenotypes, haplotype characterisation, and analysis of the population structure, so that genetic association studies preserve their quality as one of the tools more powerful to a practical approach.9
Table 1. Variables that influence sample size assessment
Table 2. Alternatives to face multiplicity
Table 3. Type of statistical analysis employed
Table 4. Statistical power and study replicability
Figure 1.