Dent's disease type 1 (DD1) is a rare X-linked hereditary pathology caused by CLCN5 mutations that is characterized mainly by proximal tubule dysfunction, hypercalciuria, nephrolithiasis/nephrocalcinosis, progressive chronic kidney disease, and low-weight proteinuria, the molecular hallmark of the disease. Currently, there is no specific curative treatment, only symptomatic and does not prevent the progression of the disease. In this study we have isolated and characterized urinary extracellular vesicles (uEVs) enriched in exosomes that will allow us to identify biomarkers associated with DD1 progression and a better understanding of the pathophysiological bases of the disease.
Materials and methodsThrough a national call from the Spanish Society of Nephrology (SEN) and the Spanish Society of Pediatric Nephrology (AENP), urine samples were obtained from patients and controls from different Spanish hospitals, which were processed to obtain the uEVS. The data of these patients were provided by the respective nephrologists and/or extracted from the RENALTUBE registry. The uEVs were isolated by ultracentrifugation, morphologically characterized and their protein and microRNA content extracted.
Results25 patients and 10 controls were recruited, from which the urine was processed to isolate the uEVs. Our results showed that the relative concentration of uEVs/mL is lower in patients compared to controls (0.26 × 106 uEVs/mL vs 1.19 × 106 uEVs/mL, p < 0.01). In addition, the uEVs of the patients were found to be significantly larger than those of the control subjects (mean diameter: 187.8 nm vs 143.6 nm, p < 0.01). Finally, our data demonstrated that RNA had been correctly extracted from both patient and control exosomes.
ConclusionsIn this work we describe the isolation and characterization of uEVs from patients with Dent 1 disease and healthy controls, that shall be useful for the subsequent study of differentially expressed cargo molecules in this pathology.
La enfermedad de Dent tipo 1 (DD1) es una patología hereditaria rara ligada al cromosoma X causado por mutaciones en el CLCN5 que se caracteriza principalmente por una disfunción del túbulo proximal, hipercalciuria, nefrolitiasis/nefrocalcinosis, enfermedad renal crónica progresiva y proteinuria de bajo peso molecular, rasgo distintivo de la enfermedad. Actualmente, no existe un tratamiento curativo específico, únicamente sintomático y no previene la progresión de la enfermedad. En este estudio hemos aislado y caracterizado las vesículas extracelulares urinarias (uEVs) enriquecidas en exosomas que nos permitirán identificar biomarcadores asociados a la progresión de DD1 y una mejor comprensión de las bases fisiopatológicas de la misma.
Materiales y métodosA través de una convocatoria nacional de la Sociedad Española de Nefrología (SEN) y la Sociedad Española de Nefrología Pediátrica (AENP), se obtuvieron orinas de pacientes y controles de distintos hospitales españoles, las cuales se procesaron para obtener los uEVS. Los datos de estos pacientes fueron proporcionados por los respectivos nefrólogos y/o extraídos del registro RENALTUBE. Los uEVs se aislaron mediante ultracentrifugación, fueron caracterizados morfológicamente y se extrajo su contenido de proteína y microRNA.
ResultadosSe reclutaron 25 pacientes y 10 controles, de los cuales se procesaron las orinas para aislar los uEVs. Nuestros resultados mostraron que la concentración relativa de uEVs/mL es menor en pacientes comparado con controles (0,26 × 106 uEVs/mL vs 1,19 × 106 uEVs/mL, p < 0.01). Además, se vio que los uEVs de los pacientes eran significativamente más grandes que los de los sujetos control (diámetro medio: 187,8 nm vs 143,6 nm, p < 0.01). Finalmente, nuestros datos demostraron que se había extraído correctamente RNA tanto de los exosomas de pacientes como de controles.
ConclusionesEn este trabajo describimos el aislamiento y caracterización de uEVs de pacientes de la enfermedad de Dent 1 y controles sanos, útiles para el posterior estudio de moléculas cargo diferencialmente expresadas en esta patología.
Dent's disease type 1 (DD1) is a rare hereditary disease first described in 1964,1 with about 400 families described so far. It is characterized by hypercalciuria and low molecular weight proteinuria, which is the hallmark of the disease.2 Patients may develop nephrocalcinosis, kidney stones or chronic kidney disease.3 In DD1, vesicle trafficking and intracellular transport are disrupted due to mutations in the electrogenic antiporter 2Cl−/H+ ClC-5 encoded by the CLCN5 gene4,5 (OMIM # 300009). CLCN5 is located on chromosome Xp11.22 and the most common isoform contains 746 amino acids.6 ClC-5 controls acidification and recycling of endosomal compartments, is predominantly expressed in renal proximal tubule cells and it is localized to in endosomes and the apical membrane,6 although it has also been described in human podocytes.6 Inactivation by mutations in CLCN5 results in severe low molecular weight proteinuria due to defective endocytic uptake in renal proximal tubule cells, which has been associated with the disappearance of megalin and cubilin in the brush border membranes of these renal cells.5 Although DD1 has been considered a tubular disease, cases with lesions of focal glomerulosclerosis have also been described in renal biopsies.7
The DD1 phenotype can be confused with that of other related inherited disorders that also lead to chronic kidney disease, making diagnosis difficult. There is considerable intra-familial variability in the clinical expression and progression of the disease, with no clear genotype-phenotype correlation.8–10 Currently, there is no specific treatment or clinical trials of this disease, so the care of these patients is focused on the treatment of hypercalcuria and the prevention of nephrolithiasis.3
To further understand the pathophysiological basis of DD1, as well as to obtain new biomarkers of disease progression, our group conducted a study based on obtaining exosome-enriched urinary extracellular vesicles (uEVs) from which disease-associated mRNA profiles can be obtained. Exosomes appear to be derived from each of the epithelial cell types facing the renal tubule lumen11 and thus may provide valuable information for monitoring physiological and pathological changes throughout the nephron using a noninvasive procedure such as collection and analysis of the urine.12
In this work, we describe the methodology used to obtain urinary vesicles and their characterization in patients with DD1 and in controls.
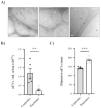
Materials and methodsPatientsUrine samples were collected from 25 male DD1 patients with confirmed genetic diagnosis and from 10 healthy donors of the same age and sex from different hospitals in Spain for processing at the Hospital Universitari Vall d'Hebron (Barcelona, Spain) (Fig. 1A). The study was approved by the Vall d'Hebron Hospital Ethics Committee (PR(AG)149/2020/ 314/C/2020). The estimated glomerular filtration rate (eGFR) was calculated using the CKD-EPI formula.13
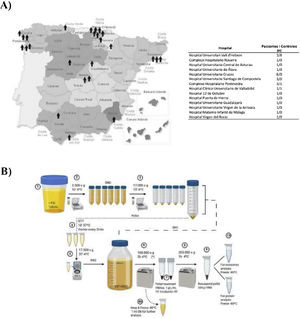
Isolation of exosomesIsolation of uEV was performed according to the protocol previously described by our group.14 This process is shown in the scheme shown in Fig. 1B.
Nanoparticle tracking analysisThe quantification and distribution of the uEV measurement was analyzed using the nanoparticle tracking analysis (NTA) technique (NanoSight NS300, Malvern Instruments, UK). This equipment uses light scattering and Brownian motion to obtain the dimensions and concentration of suspended particles.15,16 These parameters were recorded with a high sensitivity camera and the images obtained were analyzed with NTA v3.1 software (NanoSight Ltd., Malvern Instruments, UK) at the Institut de Ciència de Materials de Barcelona (ICMAB-CSIC), Universitat Autònoma de Barcelona (Spain).
Cryogenic transmission electron microscopyCryogenic transmission electron microscopy (Cryo-TEM) was performed at the Microscopy Center of the Universitat Autònoma de Barcelona. 10 μL of uEV dsiluted in 1x PBS were applied on Formvar-Carbon EM grids and frozen in liquid ethanol cooled to −179 °C. Samples were analyzed on a Jeol JEM 2011 transmission electron microscope at an acceleration voltage of 200 kV.
Extraction of exosomal proteinsSuspended uEVs were incubated with 1:1 lysis buffer (Tris−HCl 100 mM pH 7.5, EDTA 2 mM, pH 8, NaCl 300 mM, SDS 0.2%, NP-40.2%, sodium deoxycholate 0.5%, PIC 1:200, NaF 1 mM, Na 3VO 1 mM) at 4 °C under constant rotation for 1 h. Then, samples were sonicated at maximum amplitude for 5 cycles of 5 s, and then centrifuged at 13,000×g for 15 min at 4 °C. Supernatants were collected and stored at −20 °C. The uEV lysates were processed and analyzed by western blot.
RNA extraction and quantificationFor RNA extraction from previously characterized uEV lysates we used the miRNeasy Mini kit (Qiagen). Quantification of RNA levels was performed using the BioAnalyzer 2100 in combination with the RNA 6000 Pico LabChip kit.
Statistical analysisResults are presented as mean ± standard error of the mean (SEM). Student's t-test (2-tailed) was used for statistical analysis. A p value below 0.05 was considered to indicate statistically significant differences. Statistical analyses were performed with commercial software (GraphPad Prism, version 9 for MacOS, GraphPad Software, La Jolla California, USA).
ResultsStudy populationDD1 is an ultra-rare disease with a low number of diagnosed patients. To obtain sufficient patients for this study, a national call was made through the Spanish Society of Nephrology (SEN) and the Spanish Society of Pediatric Nephrology (AENP). A total of 25 patients with confirmed mutations in the CLCN5 gene from 14 hospitals in Spain participated in this study, from whom samples and clinical data were collected (Fig. 1A). Ten age- and race-matched healthy controls were also included (all individuals studied were male). Patient demographic and clinical data confirmed that there were no significant differences in age between the two groups (p = 0.67) but showed that 44% of them (p = 0.012) had reduced renal function (GFRe <90 mL/min/1.73 m2) and 52% had nephrocalcinosis (p = 0.0052) (Table 1). There were no significant differences in the rest of the parameters between patients and controls.
Clinical characteristics of the cohort (controls and patients).
| Patients (n = 25). | Controls (n = 10) | p | |
|---|---|---|---|
| Age at last follow-up, median years, [IQR]. | 11.5 [4.75–19] | 10 [8.5–25] | 0.67 |
| Sex, male n (%) | 25 (100) | 10 (100) | >0.99 |
| Chronic kidney disease | |||
| Without renal failure (>90 mL/min/1.73 m2) | 12 (52.2) | 10 (100) | 0.0129 |
| Stage 2 (60–89 mL/min/1.73 m2) | 5 (21.7) | 0 (0) | |
| Stage 3 (30–59 mL/min/1.73 m2) | 3 (13) | 0 (0) | |
| Stage 4 (15–29 mL/min/1.73 m2) | 3 (13) | 0 (0) | |
| Stage 5 (<15 mL/min/1.73 m2) or RRT | 0 (0) | 0 (0) | |
| Nephrocalcinosis, yes n (%) | 13 (52) | 0 (0) | 0.0052 |
| Lithiasis, yes n (%) | 4 (16) | 0 (0) | 0.303 |
| Incomplete Fanconi syndrome (glycosuria or aminoaciduria) n (%) | 4 (16) | 0 (0) | 0.303 |
| Rickets, yes n (%) | 3 (12) | 0 (0) | 0.542 |
Bold: significant difference.
IQR: interquartile range; RRT: renal replacement therapies.
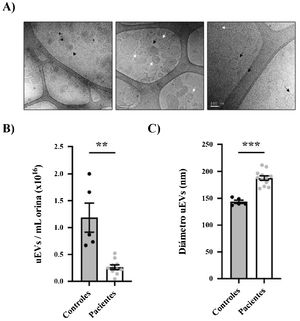
Urine samples from patients and controls were processed to obtain uEVs as previously described (Fig. 1B).14 Exosome-enriched uEVs obtained from patients and controls were morphologically characterized using the cryo-TEM technique, which allows direct visualization of vesicles without the addition of heavy metals or fixatives, that could cause artifacts. Our results revealed single-, double- or multilayered vesicles as well as uEVs of different sizes without obvious impurities (Fig. 2A).
Morphological characterization of uEVs in patients and controls. (A) Details of representative samples with different scales (500 nm, 200 nm and 100 nm from left to right) showing single (white arrow), double (continuous black arrow) or multilayered (dashed black arrow) vesicles. (B) Quantification of uEV concentration per mL of urine in controls (black circles) or patients (gray circles). Each point represents one subject. The mean ± SEM is also shown in the graph. (C) Quantification of uEV diameter of controls (black circles) and patients (gray circles). Each point represents the mean uEV diameter for a single subject. The graph also shows the mean ± SEM.
CD81: differentiation group 81; GRP78: glucose regulatory protein 78; HepG2: hepatocellular carcinoma cell line G2; uEV: urinary exosome-like vesicles; SN: supernatant.
** p < 0.01.
***p < 0.001.
The size distribution and relative uEV concentration were evaluated by nanoparticle tracking analysis (NTA) in randomly selected samples (n = 12 patients, n = 5 controls).15,16 It was very interesting to note that the relative concentration of uEV/mL was significantly lower in patients than in controls (0.26 ± 0.04 × 1016 vs. 1.19 ± 0.27 × 1016 uEV/mL; p = 0.0004) (Fig. 2B). As seen in Fig. 2C, most of the detected vesicles were less than 200 nm in diameter, as expected for exosomes. Curiously, exosomes from patients were significantly larger than those from control subjects (mean diameter 187.8 ± 4.053 vs. 143.6 ± 2.994 nm, respectively; p < 0.0001) (Fig. 2C).
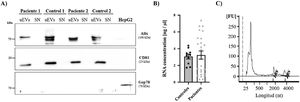
Finally, the expression of two exosome-associated markers (ALIX and CD81)17 was evaluated by western blotting to confirm the presence of urinary exosomes (Fig. 3A). Both markers were detected in all exosome samples (both patients and controls) and were absent in the supernatant of these same samples, which were used as a negative control to confirm the absence of contamination by cellular debris. In addition, we used an endoplasmic reticulum marker (GRP78) to demonstrate the absence of intracellular structures in the exosome suspensions.
RNA and protein obtained from uEVs. (A) Alix and CD81 (exosome markers) levels were analyzed by western blot in patient and control samples. HepG2 cell line lysate was used as a positive control for the detection of Grp78, a reticulum marker. (B) RNA concentration (ng/μL) in samples from controls (black circles) and patients (gray circles). Each point represents a single subject. The graph also shows the mean ± SEM. (C) Representative electropherogram showing good integrity of RNA obtained from uEVs. Peaks corresponding to 18S or 28S ribosomal RNA are marked on the graph.
FU: arbitrary fluorescence units; nt: nucleotides.
RNA extracted from the uEVs was quantified using the BioAnalyzer 2100. Total RNA quantification showed no differences between controls and patients (Fig. 3B) and similar electropherograms were obtained in all samples, as well as good RNA integrity in all cases. As exhibited in the example in Fig. 3C, a high peak corresponding to 250 nt RNA and two lower but larger peaks corresponding to 18S or 28S ribosomal RNA were observed (Fig. 3C).
DiscussionThe aim of this study was to identify and select Spanish patients with a confirmed genetic diagnosis of DD1, as well as sex and age matching healthy controls for the isolation and characterization of exosome-enriched uEVs. In addition, RNA has been extracted from the uEVs to further study the differential expression profile between patients and controls foridentification of diagnostic and prognostic biomarkers, and possible therapeutic targets against DD1. Indeed, urine sample analysis is irreplaceable as a non-invasive method for the diagnosis and monitoring of some diseases, particularly renal diseases. In contrast to tissue biopsy, an invasive and complicated procedure that allows only a partial sample of the organ to be obtained, exosome-enriched uEVs provide a complete representation of the entire urinary system and their study is attractive in the field of biomarker discovery. The potential use of uEVs as diagnostic and prognostic markers in clinical studies has been evaluated, as well as their contribution in pathophysiological processes using experimental models.18–20
Isolation of exosomesUrine samples from patients from all over Spain were frozen, after an initial centrifugation, until processing. Although some authors have pointed out that direct freezing at −80 °C could better preserve exosomes,21 others have shown that exosomes and their charges are stable in urine under different storage conditions, by obtaining comparable results with freshly processed samples or after repeated freeze-thaw cycles.22 For uEV isolation, the differential centrifugation method was preferred to other techniques.23 No previous data have been published on uEV in DD1 patients and, therefore, the efficacy of exosome isolation per urine sample in these polyuric patients, who, in addition, have impaired endocytosis and endolysosomal pathways, both of which are involved in exosome formation, was unknown.
This technique has been widely used in our laboratory and has already proven effective in the isolation of exosomes to study another rare renal tubulopathy with polyuria.14
Differences in number and size of exosomesQuantification of uEVs by NTA revealed a substantial concentration, although highly variable concentrations between individual samples. This variation was independent of the glomerular filtrate or the degree of proteinuria, but other factors could play a role, such as the amount of Tamm Horsfall protein. Interestingly, patients were observed to have almost five times lower uEV/mL urine concentration than controls. This fact may be related to the polyuria observed in people with DD1.
However, we also observed significant differences in the size of the exosomes of the patients compared to those of the controls, as mentioned above. One possible hypothesis would be that some defects in the endolysosomal pathway, resulting from ClC-5 dysfunction, produced by an unknown mechanism the release of larger vesicles in DD1 patients than in controls. In fact, this change in size could also explain the reduction in the number of particles between patients and controls: fewer exosomes are released, but they are larger.
The tubular dysfunction that occurs in Dent's disease could have an impact on the production and release of exosomes, since their biogenesis begins in the endosomal system, where early endosomes mature into late endosomes or multivesicular bodies.24 The electrogenic antiporter (2Cl−/H+) ClC-5 is localized in early endosomes and has a very relevant role in vesicle trafficking and intracellular transport,4,5 which is altered when mutations occur in the CLCN5 gene, as occurs in DD1. Likewise, it has been pointed out that the fate of multivesicular bodies, either their degradation or exosome secretion, depends on cellular homeostasis, which is probably altered in patients with Dent's disease. Multivesicular bodies may be directed to lysosomes, where their contents are degraded, or transported to the plasma membrane for the release of exosomes.25 Little is known about the molecular and cellular mechanisms that regulate this balance and the role of ClC-5 in these processes remains to be determined.
In other diseases, such as neurodegenerative dementias, a reduction in the concentration of extracellular vesicles in plasma has also been seen, with an increase in their size.26 However, no mechanism explaining this change has yet been reported in the literature. On the other hand, Paulaitis et al. have characterized exosome size distributions based on the dynamic scale of multivesicular body growth in the exosome biogenesis pathway.27 These authors find statistically significant differences in the scaling exponents characterizing exosome size distributions in serum samples from cancer patients compared to those from healthy donors. The authors consider that such changes could reflect alterations in the membrane composition of exosomes and propose that the scaling exponent could be considered as a biophysical biomarker comparable to and complementary to biochemical biomarkers of exosomes, such as specific membrane proteins such as tetraspanins.
ConclusionsIn this study we have isolated and characterized for the first time exosomes from DD1 patients. In fact, morphological characterization of these exosomes has shown that there is a decrease in their concentration in urine together with an increase in their size. We do not have an explanation for these findings, however an in-depth study of this observation should allow us to explain the possible role of ClC-5 in the biogenesis of exosomes. Finally, these results will allow us to study microRNAs from urinary exosomes to find possible biomarkers and therapeutic targets for Dent's disease.
Authors' contributionAM conceived and designed the project. CB and MD carried out the experimental part. AM, CB and GCR analyzed and interpreted the data. AM prepared the draft article. CB, MD, CM, GCR, GA and AM participated in revising and writing the final manuscript.
FinancingThis work has been mainly funded by the SENEFRO foundation (SEN 2019 to AM), by ASDENT and by grants from the Ministry of Science and Innovation (SAF201789989 to AM) and from the Renal Research Network REDinREN (12/0021/0013). The Renal Pathophysiology Group has the Mention of Quality of the Generalitat de Catalunya (2021 SGR 01600).
Conflict of interestThe authors declare no conflicts of interest.
We thank the patient association ASDENT (www.asdent.es) for their continued support; without them, this project would not have been possible. We also thank all members of the Renal Pathophysiology Group for valuable discussions. This work reflects only the authors' points of view.
MD was hired thanks to the generous contribution of ASDENT and CB was awarded a PhD4MD fellowship for physicians from VHIR and CRG.