Diabetic Nephropathy (DN) is a major complication of Type 2 Diabetes Mellitus (T2DM) with high morbidity rates worldwide.
ObjectiveTo determine the association of PPARγ rs1801282 polymorphism in T2DM and DN in south Indian population.
MethodsWe have conducted a case–control study to test the association of rs1801282 polymorphism with T2DM and DN in 424 subjects (DN=128; T2DM=148 and controls=148) belonging to the south Indian population using ARMS-PCR and Sanger sequencing method. Further, a meta-analysis was performed for rs1801282 polymorphism from the published literature retrieved from various electronic databases to determine the susceptibility among T2DM and DN across various ethnic populations under five genetic models.
ResultsThe genotyping of rs1801282 polymorphism showed significant (p-value<0.05) association with DN and T2DM compared to controls. In the meta-analysis, no significant association (p-value>0.05) was noticed for rs1801282 with DN vs. controls in homozygote, heterozygote, allelic, recessive and dominant genetic models. However, a significant association was observed between rs1801282 SNP and T2DM under heterozygote (Jj vs JJ) genetic model with OR=0.56, (95%CI [0.43–0.74]), p≤0.0001 of Asian and Caucasian populations.
ConclusionOverall analysis suggests that the rs1801282 polymorphism might be associated with DN and T2DM. More case–control studies on the PPARγ gene with a larger sample size including all the confounding factors are required to corroborate the findings from this meta-analysis.
La nefropatía diabética (ND) es una complicación importante de la diabetes mellitus de tipo 2 (DMT2) con altas tasas de morbilidad mundial.
ObjetivoDeterminar la asociación del polimorfismo rs1801282 de PPARγ en la DMT2 y la ND en la población del sur de India.
MétodosHemos llevado a cabo un estudio de casos y controles para analizar la asociación del polimorfismo rs1801282 con la DMT2 y la ND en 424 sujetos (ND=128; DMT2=148 y controles=148) pertenecientes a la población del sur de India mediante RCP-ARMS y método de secuenciación de Sanger. Además, se realizó un metaanálisis para el polimorfismo de rs1801282 a partir de la literatura publicada en varias bases de datos electrónicas para determinar la sensibilidad entre la DMT2 y la ND en varias poblaciones étnicas con 5 modelos genéticos.
ResultadosEl genotipado de polimorfismo rs1801282 demostró una asociación significativa (valor de p<0,05) con la ND y la DMT2 en comparación con los controles. En el metaanálisis no se observó asociación significativa (valor de p>0,05) de rs1801282 con la ND frente a los controles en modelos genéticos homocigóticos, heterocigóticos, alélicos, recesivos y dominantes. Sin embargo, se observó una asociación significativa entre el polimorfismo de nucleótido único (PNU) rs1801282 SNP y la DMT2 en el modelo genético heterocigótico (Jj frente a JJ) con OR=0,56, (IC del 95%: 0,43-0,74; p≤0,0001 de poblaciones asiáticas y caucásicas.
ConclusiónEl análisis general sugiere que el polimorfismo rs1801282 puede asociarse a ND y a DMT2. Se precisan más estudios de casos y controles sobre el gen PPARγ con un tamaño de la muestra mayor que incluya todos los factores de confusión para corroborar los resultados de este metaanálisis.
Diabetes Mellitus (T2DM) is a complex metabolic disorder characterized by abnormal lipid, protein metabolism, and hyperglycemia. T2DM is considered as serious health problem globally and ranks as the world's sixth-leading factor causing death.1 Diabetic Nephropathy is one of the major complication of T2DM and the primary cause of the end-stage renal disease (ESRD), with a high morbidity rate from 25 to 40% globally.2 Abnormal glycaemic control and diabetes duration are considered as the major risk factors for DN. The pathogenesis and advancement of T2DM to DN is not well established, previous epidemiological and clinical studies have demonstrated the role of hyperlipidemia in the glomerular injury through the initiation and activation of multiple signaling pathways.3 A meta-analysis was performed in the year 2011 to evaluate the genetic associations with DN from various ethnic populations which revealed 21 significantly associated genes such as APOC1, ACE, AKR1B1, APOE, CHN2, EPO, ‘GREM1, NOS3, HSPG2, FRMD3, CPVL, VEGFA, CARS, and UNC13B.4 In addition, several Genome-Wide Association studies have documented 56 genetic loci associated with T2DM susceptibility. Also, few single nucleotide polymorphisms (SNPs) were reported in candidate genes such as FTO, MC4R, PPARγ, and ADIPOQ with T2DM.5,6
Peroxisome Proliferator-Activated Receptor-Gamma (PPARγ) gene belongs to the nuclear hormone receptor subfamily which controls the expression of genes involved in insulin sensitivity, lipogenesis, adipocyte differentiation, inflammation, and metabolic syndrome.7 PPARγ gene is localized at 3p25.2 spanning 9 exons encodes 505 amino acids. The PPARs mainly comprise nuclear fatty acid receptors, which have a hydrophobic ligand-binding pocket and a zinc finger DNA binding motif (type-II).8 To date, numerous case–control studies have been conducted to identify the possible relationship between PPARγ gene polymorphisms with the risk of T2DM and DN in various ethnic populations.9 The most common variant located in exon-2 of PPARγ rs1801282 (Proline-12-Alanine), leading to the substitution of cytosine to guanosine at 34th nucleotide position. This substitution leads to a change in the structure of PPARG protein, which in turn decreases the binding effect of target genes and thereby reducing transcriptional activity.10 In addition, studies have indicated that rs1801282 polymorphism is found involved in reducing the albuminuria risk and insulin resistance among DM patients.11 Among worldwide population, Indians belong to the Asian ethnic background, considered as a high-risk population for T2DM. It has been well documented that Asian Indians have a high waist-hip ratio, greater insulin resistance, increased susceptibility to diabetes and posses the risk of coronary heart disease (CHD) compared with Europeans or the Caucasoids.12 We conducted a case–control study among the south Indian population belong to Asian ethnic background to determine the rs1801282 association in DN, T2DM, compared to healthy controls, Further, the resulting genotypic frequencies of rs1801282 polymorphism were analyzed with the previously published literature based on the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) criteria to confirm its significance across various populations.
MethodsSample characteristicsThe samples (diabetic nephropathy, type-2 diabetes mellitus, and healthy controls) for this study were recruited from the subjects attending Department of General Medicine, Chettinad Hospital and Research Institute (CHRI), Tamil Nadu, India, from January to June 2016. The selection criteria for DN subjects by clinical examination including the routine biochemical assessment such as excreting urine albuminuria (>300mg/L) and serum creatinine (>1mg/dL) were considered and histopathological investigation. The T2DM subjects were diagnosed based on the criteria defined by the World Health Organization (WHO).13 The controls were selected by following the similar clinical and biochemical examination as done for DN and T2DM. All study participant were age-matched, belonging to South India, Asian ethnicity. The current study protocols were reviewed and approved by the Institutional Human Ethics Committee (03/IHEC/3-16) of the Chettinad Academy of Research and Education. Informed consent was obtained from the participants before the sampling process. Likewise, HIV positive patients, Juvenile diabetic subjects, physically challenged and subjects with cardiac problems were excluded. The demographic data and baseline clinical characteristics from each study subject including gender, age, Body Mass Index (BMI), the onset of disease, symptoms of the disease, family history, diet, and physical activity were recorded at the time sample collection.
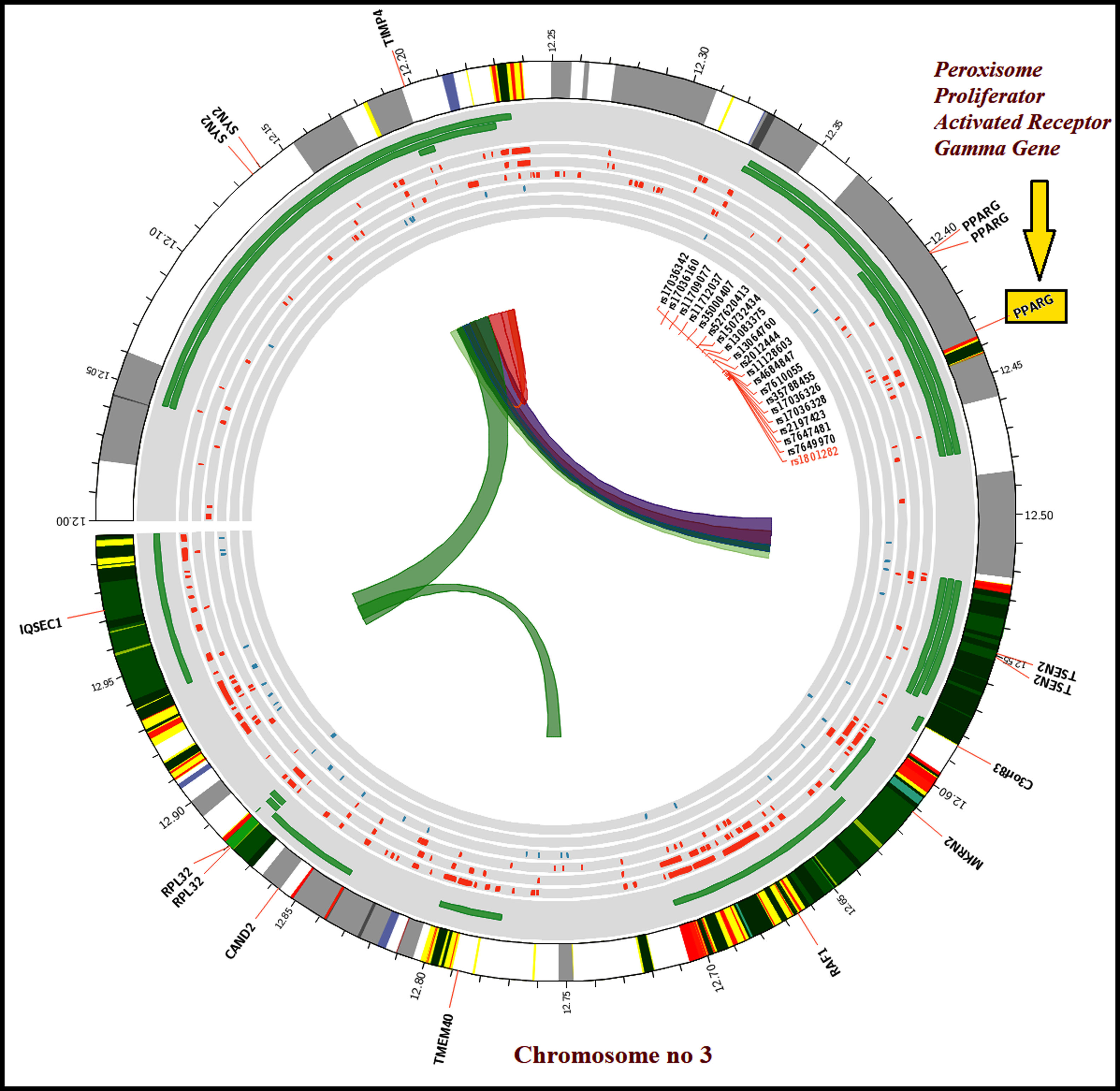
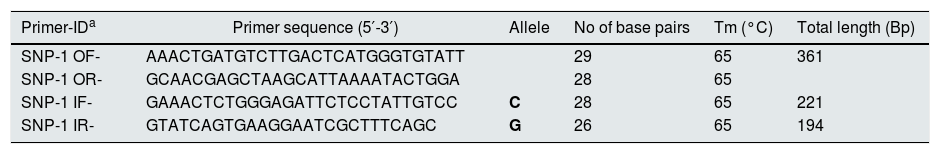
Genetic analysis of rs1801282 polymorphismVenous blood (3ml) was collected in EDTA coated tube (BD, USA) from the participants to isolate Genomic DNA using the phenol-chloroform method with minor modifications.14 The genetic analysis of rs1801282 was performed by Amplification Refractory Mutation System-Polymerase Chain Reaction (ARMS-PCR) using allele-specific primers (Table 1).15 For each PCR reaction, a 10μL reaction mix was setup containing 25ng DNA, 1U TAQ Brazilian Origin, 0.3mmol of each dNTP, 12pmol/μL of each primer. The ARMS-PCR program was performed using an ABI Veriti® Thermal Cycler (California, USA). The reaction condition for ARMS-PCR was set as initial denaturation (94°C, 5min), denaturation (94°C, 45s), annealing (68°C, 45s), elongation (72°C, 45s) and final elongation at 72°C for 5min. The amplicons were visualized on 1.6% agarose gel together with 100bp DNA Ladder (Dye Plus, Cat no: 3422A, Takara Bio). Finally, the identified genotypes were further confirmed in the randomly selected subset of overall samples (DN=12; T2DM =10; controls=08) using Sanger based sequencing (Applied Biosystems 3130, USA). Additionally, to determine the chromosomal interactions among the SNP variants, we used 3DSNP (http://www.cbportal.org/3dsnp/index.html) software package to generate Circos (circular) plots based on r2 values for visualizing the genomic data.
Designed primers for PPARγ (rs1801282) genotyping.
| Primer-IDa | Primer sequence (5′-3′) | Allele | No of base pairs | Tm (°C) | Total length (Bp) |
|---|---|---|---|---|---|
| SNP-1 OF- | AAACTGATGTCTTGACTCATGGGTGTATT | 29 | 65 | 361 | |
| SNP-1 OR- | GCAACGAGCTAAGCATTAAAATACTGGA | 28 | 65 | ||
| SNP-1 IF- | GAAACTCTGGGAGATTCTCCTATTGTCC | C | 28 | 65 | 221 |
| SNP-1 IR- | GTATCAGTGAAGGAATCGCTTTCAGC | G | 26 | 65 | 194 |
a IF-inner forward, IR-inner reverse, OF-outer forward, OR- outer reverse.
The statistical analysis for case–control study was executed by SPSS V.21 (IBM-Analytics, USA) software. The allelic and genotypic frequencies of rs1801282 polymorphism in DN, T2DM and healthy controls were calculated using the Pearson Chi-square test. To determine the distribution of SNP in controls, Hardy-Weinberg Equilibrium (HWE value>0.05) was used. Additionally, their association of polymorphism was analyzed by determining the odds ratio (OR), confidence intervals (95% CIs) in dominant (Jj+jj vs. Jj) (J-major, j-minor allele) and recessive (jj vs. JJ+Jj) genetic models.
Literature search strategy for Meta-analysisAn extensive literature to select the studies published between January 1991 to December 2017, that discuss the association between rs1801282 polymorphism with DN and T2DM from the database of MEDLINE, PubMed, Cochrane Library, EMBASE, and Google Scholar. The multiple key terms such as “Diabetic Nephropathy” “Type 2 Diabetes Mellitus”, “DN”, “T2DM”, “Peroxisome proliferator-activated receptor gamma gene”, “PPARγ gene”, “Polymorphism”, “rs1801282” were used to retrieve the relevant articles published in English Language for the meta-analysis with Diabetic Nephropathy vs. Type 2 Diabetes Mellitus. Similarly, multiple keywords such as “Type 2 Diabetes Mellitus”, “T2DM”, “Peroxisome proliferator-activated receptor gamma gene”, “PPARγ gene”, “Genetic association studies”, “SNP”, “rs1801282” were used to perform the similar meta-analysis between T2DM vs. controls.
Data extraction for meta-analysisStudies included in this meta-analysis (Diabetic Nephropathy vs. Type 2 Diabetes Mellitus; Type 2 Diabetes Mellitus vs. controls) were essential to meet the following criteria's: first, the association of PPARγ gene and rs1801282 polymorphism with DN and T2DM, T2DM and controls should be assessed. Second, study should be cases–controls (two groups). Third, the articles should provide adequate data to calculate allele and genotype frequencies for the meta-analysis. The information from the collected articles was extracted by two authors [AH and PA], any discrepancy between the authors was resolved by a group discussion (IR, SSJ, and RK). The study characteristics such as first author name, year of publication, country, ethnicity, the source of DNA, the sample size for cases-controls, genotypic frequency and genotyping method were extracted.
Quality assessment and meta-analysis for rs1801282 polymorphismThe methodological quality assessment of each study was evaluated by two independent variables such as the Hardy-Weinberg Equilibrium (HWE)16 and the Newcastle Ottawa Scale (NOS).17 The genotype distribution of included studies among the controls should follow HWE p-value above 0.05. The NOS grading system relies on three main factors such as selection, comparability, and exposure. The studies score up to a maximum of nine points, six or above were considered in this meta-analysis. In the meta-analysis, rs1801282 SNP with risk of DN vs. T2DM and T2DM vs. controls, were performed to calculate the pooled odds ratios (OR) and 95% confidence interval (CI) with (p-value<0.05) under allelic (j vs. J) (J-major, j-minor allele), homozygote (jj vs. JJ), heterozygote (Jj vs. JJ), dominant (Jj+jj vs. JJ) and recessive (jj vs. JJ+Jj) genetic models. The heterogeneity between the included studies in this meta-analysis was assessed by the I2 test18 and Q-statistics. Based on the results obtained from heterogeneity (I2<50), fixed effect (Mantel-Haenszel's)19 method was used, if not, the random-effect method (DerSimonian and Laird's) was applied. The publication bias for meta-analysis was assessed by Begg's funnel plot and Egger's test.20 Leave-one-out method was executed to validate the consistency of our results.21 The meta-analysis was performed using Revman V.5.0 (Cochrane Collaboration, UK) and STATA V.12.0 (Stata Corp, USA).
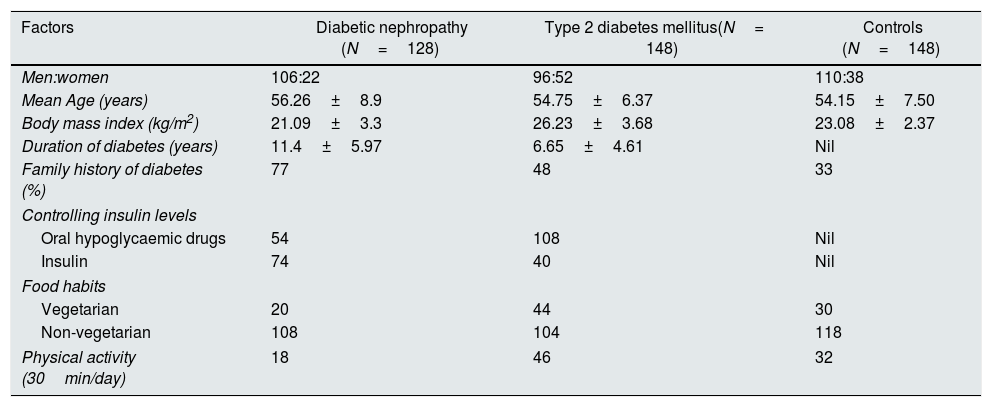
ResultsCharacteristics of study subjects in case–control studyThe baseline clinical characteristics of the study participants (DN=128; T2DM=148 and controls=148) were illustrated in Table 2. Among the samples enrolled in the study, their average±SD of age in DN, T2DM, and control groups were 56.26±8.9, 54.75±6.37 and 54.15±7.50 years respectively. Similarly, the body mass index (BMI) were (21.09±3.3), (26.23±3.68) and (23.08±2.37) for DN, T2DM and control.
Demographic and base-line clinical characteristics of DN, T2DMand Controls.
| Factors | Diabetic nephropathy (N=128) | Type 2 diabetes mellitus(N= 148) | Controls (N=148) |
|---|---|---|---|
| Men:women | 106:22 | 96:52 | 110:38 |
| Mean Age (years) | 56.26±8.9 | 54.75±6.37 | 54.15±7.50 |
| Body mass index (kg/m2) | 21.09±3.3 | 26.23±3.68 | 23.08±2.37 |
| Duration of diabetes (years) | 11.4±5.97 | 6.65±4.61 | Nil |
| Family history of diabetes (%) | 77 | 48 | 33 |
| Controlling insulin levels | |||
| Oral hypoglycaemic drugs | 54 | 108 | Nil |
| Insulin | 74 | 40 | Nil |
| Food habits | |||
| Vegetarian | 20 | 44 | 30 |
| Non-vegetarian | 108 | 104 | 118 |
| Physical activity (30min/day) | 18 | 46 | 32 |
Data are presented as mean±SD.
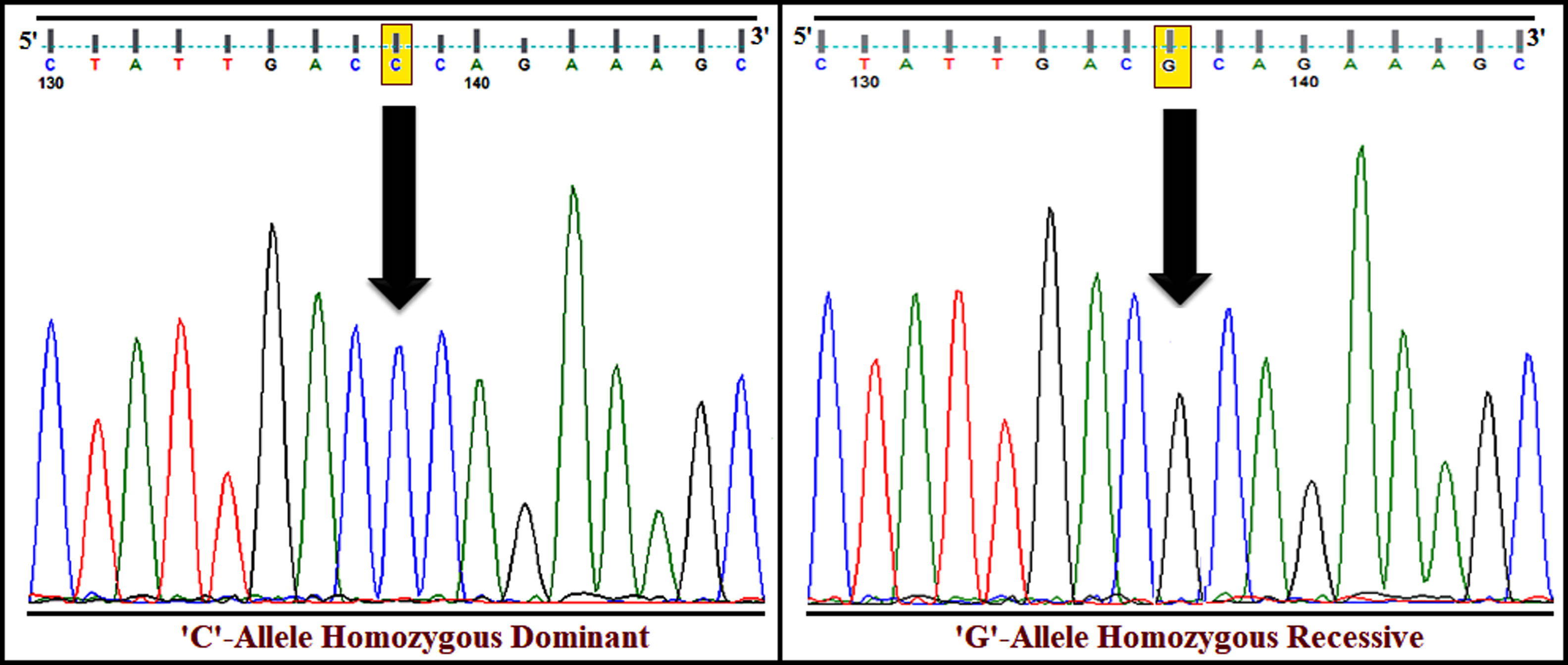
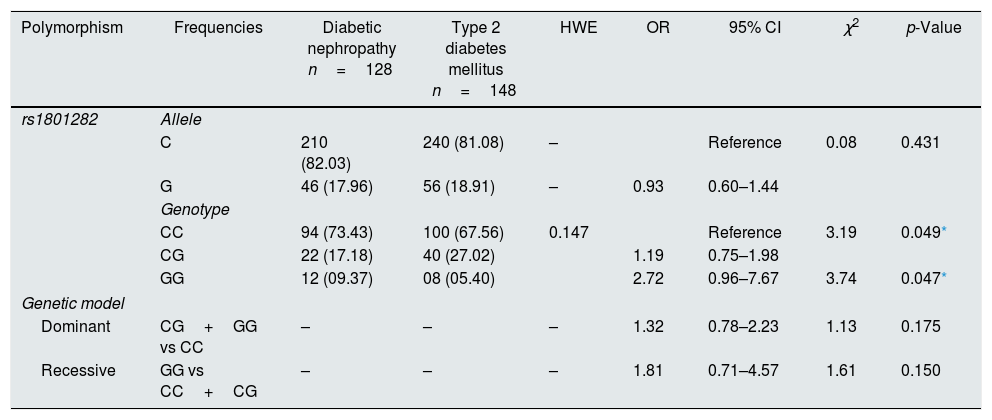
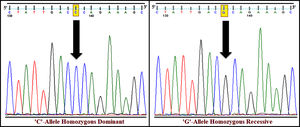
The genotyping of PPARγ gene polymorphism (rs1801282) was performed using ARMS-PCR and Sanger dideoxy sequencing (Fig. 1). The genotype frequency distributions in controls (T2DM) were in concurrence with Hardy–Weinberg Equilibrium (p>0.05). The allele and genotype frequencies of rs1801282 polymorphisms were assessed to determine the odds ratio (OR), confidence intervals (95%CIs), χ2 and p-value (Table 3). The genotype distribution of rs1801282 polymorphism was 73.43% for JJ, 17.18% for Jj, 09.37% for jj in DN cases and 67.56%, 27.02%, 5.40% in T2DM, respectively. The analysis of rs1801282 polymorphism illustrated a positive association with DN compared with T2DM (p-value≤0.05) in south India. In heterozygous (Jj) and homozygous recessive genotype (jj) model of rs1801282 showed significant risk association in DN patients compared with T2DM with OR=1.19 (95%CI [0.75–1.98]) p-value=0.049, OR=2.72 (95%CI [0.96–7.67]) p-value=0.047, respectively. Further, the analysis of dominant and recessive genetic models showed an insignificant difference between DN and T2DM for rs1801282 polymorphism.
Association of the PPARγ Gene polymorphism with DN and T2DM.
| Polymorphism | Frequencies | Diabetic nephropathy n=128 | Type 2 diabetes mellitus n=148 | HWE | OR | 95% CI | χ2 | p-Value |
|---|---|---|---|---|---|---|---|---|
| rs1801282 | Allele | |||||||
| C | 210 (82.03) | 240 (81.08) | – | Reference | 0.08 | 0.431 | ||
| G | 46 (17.96) | 56 (18.91) | – | 0.93 | 0.60–1.44 | |||
| Genotype | ||||||||
| CC | 94 (73.43) | 100 (67.56) | 0.147 | Reference | 3.19 | 0.049* | ||
| CG | 22 (17.18) | 40 (27.02) | 1.19 | 0.75–1.98 | ||||
| GG | 12 (09.37) | 08 (05.40) | 2.72 | 0.96–7.67 | 3.74 | 0.047* | ||
| Genetic model | ||||||||
| Dominant | CG+GG vs CC | – | – | – | 1.32 | 0.78–2.23 | 1.13 | 0.175 |
| Recessive | GG vs CC+CG | – | – | – | 1.81 | 0.71–4.57 | 1.61 | 0.150 |
HWE, Hardy–Weinberg equilibrium, OR, Odd's ratio; χ2, Chi-square; p value-one tailed test.
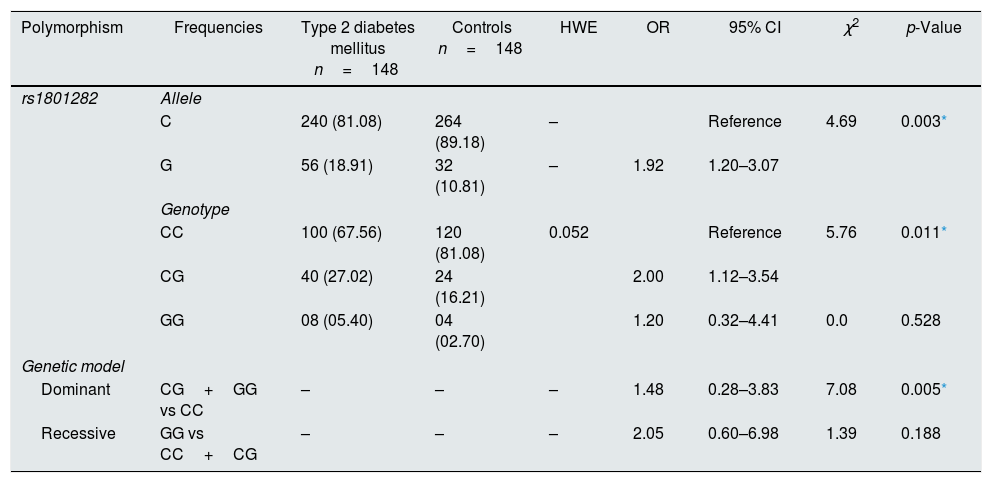
The susceptibility analysis of rs1801282 polymorphism with T2DM and controls was performed. The genotypic frequency distribution in healthy controls was in agreement with HWE (p>0.05). The allele and genotype frequencies of rs1801282 polymorphisms were used to estimate the odds ratio (OR), confidence intervals (95% CIs), χ2 and p-value (Table 4). The genotypic distribution of rs1801282 polymorphism was 67.56% for JJ, 27.02% for Jj, 05.40% for jj in T2DM and 81.08%, 16.21%, 02.70% in controls, respectively. The analysis of rs1801282 polymorphism showed a positive association with T2DM compared with controls (p-value≤0.05). The minor ‘G’ allele showed a positive association among T2DM and controls with OR=1.92 (95%CI [1.20–3.07]) p-value=0.003. In heterozygous (Jj) genotype of rs1801282 showed significant risk association among T2DM and controls with OR=2.00 (95%CI [1.12–3.54]) p-value=0.011, respectively. Likewise, the analysis of dominant genetic model showed significant association among T2DM and controls with OR=1.48 (95%CI [0.28–3.83]) p-value=0.005 for rs1801282 polymorphism.
Association of the PPARγ Gene polymorphism with T2DM and controls.
| Polymorphism | Frequencies | Type 2 diabetes mellitus n=148 | Controls n=148 | HWE | OR | 95% CI | χ2 | p-Value |
|---|---|---|---|---|---|---|---|---|
| rs1801282 | Allele | |||||||
| C | 240 (81.08) | 264 (89.18) | – | Reference | 4.69 | 0.003* | ||
| G | 56 (18.91) | 32 (10.81) | – | 1.92 | 1.20–3.07 | |||
| Genotype | ||||||||
| CC | 100 (67.56) | 120 (81.08) | 0.052 | Reference | 5.76 | 0.011* | ||
| CG | 40 (27.02) | 24 (16.21) | 2.00 | 1.12–3.54 | ||||
| GG | 08 (05.40) | 04 (02.70) | 1.20 | 0.32–4.41 | 0.0 | 0.528 | ||
| Genetic model | ||||||||
| Dominant | CG+GG vs CC | – | – | – | 1.48 | 0.28–3.83 | 7.08 | 0.005* |
| Recessive | GG vs CC+CG | – | – | – | 2.05 | 0.60–6.98 | 1.39 | 0.188 |
HWE, Hardy–Weinberg equilibrium; OR, Odd's ratio; χ2, Chi-square; p-value-one tailed test.
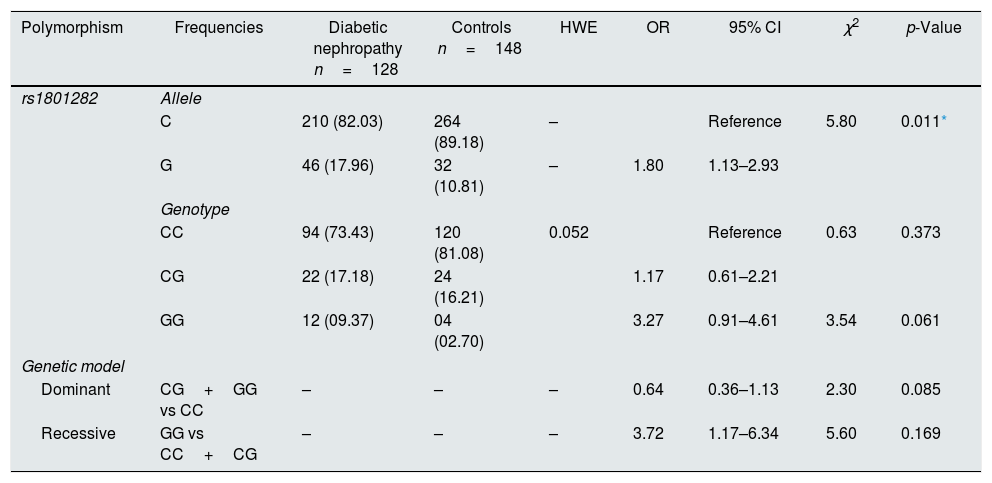
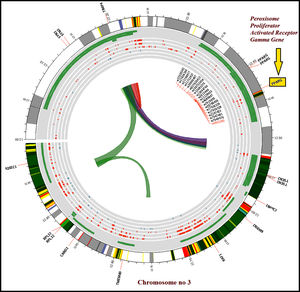
The susceptibility analysis of rs1801282 variant with DN and controls was performed (Table 5). The genotype frequency distributions in controls were in agreement with HWE (p>0.05). The genotype distribution of rs1801282 polymorphism was 73.43% for JJ, 17.18% for Jj, 09.37% for jj in DN cases and 81.08%, 16.21%, 02.70% in control, respectively. The minor ‘G’ allele showed a positive association among T2DM and control with OR=1.80 (95%CI [1.13–2.93]) p-value=0.011. However, the analysis of rs1801282 polymorphism showed a negative association between DN and control with p-value≥0.05 in the heterozygote, homozygous recessive, dominant and recessive genetic models, respectively. The nucleotide sequences of PPARγ genotypes were deposited (Accession numbers: KY547832, KY547833, KY547834, KY547835, and KY547836) in the NCBI Genbank [https://www.ncbi.nlm.nih.gov/genbank/] database. The Circos plot (outer circle to inner) illustrates the chromatin states, annotated genes, histone modifications, transcription factors, currently studied variant (rs1801282) with related SNPs (r2) and 3D chromatin interactions (Fig. 2).
Association of the PPARγ gene polymorphism with DN and controls.
| Polymorphism | Frequencies | Diabetic nephropathy n=128 | Controls n=148 | HWE | OR | 95% CI | χ2 | p-Value |
|---|---|---|---|---|---|---|---|---|
| rs1801282 | Allele | |||||||
| C | 210 (82.03) | 264 (89.18) | – | Reference | 5.80 | 0.011* | ||
| G | 46 (17.96) | 32 (10.81) | – | 1.80 | 1.13–2.93 | |||
| Genotype | ||||||||
| CC | 94 (73.43) | 120 (81.08) | 0.052 | Reference | 0.63 | 0.373 | ||
| CG | 22 (17.18) | 24 (16.21) | 1.17 | 0.61–2.21 | ||||
| GG | 12 (09.37) | 04 (02.70) | 3.27 | 0.91–4.61 | 3.54 | 0.061 | ||
| Genetic model | ||||||||
| Dominant | CG+GG vs CC | – | – | – | 0.64 | 0.36–1.13 | 2.30 | 0.085 |
| Recessive | GG vs CC+CG | – | – | – | 3.72 | 1.17–6.34 | 5.60 | 0.169 |
HWE, Hardy–Weinberg equilibrium; OR, Odd's ratio; χ2, Chi-square; p-value-one tailed test.
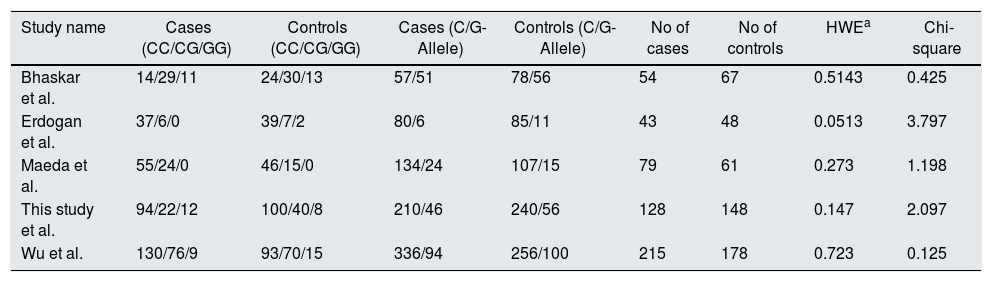
In meta-analysis, a total of 678 papers published before December 2017 was retrieved from the initial search in literature databases. The editorials, review articles, duplicates, and case reports were removed after a screening of the abstracts. Further, the studies were assessed for quality control methods such as Hardy-Weinberg Equilibrium and Newcastle Ottawa Scale. Thus, four studies22–25 were added to the meta-analysis of rs1801282 polymorphism and the characteristics of included studies in this meta-analysis were explained in supplementary Table 1. The genotypic, allelic frequencies and HWE/Chi-square values for the included studies were presented in Table 6.
Genotype and allele frequencies of PPARγ gene polymorphism with DN and T2DM.
| Study name | Cases (CC/CG/GG) | Controls (CC/CG/GG) | Cases (C/G-Allele) | Controls (C/G-Allele) | No of cases | No of controls | HWEa | Chi-square |
|---|---|---|---|---|---|---|---|---|
| Bhaskar et al. | 14/29/11 | 24/30/13 | 57/51 | 78/56 | 54 | 67 | 0.5143 | 0.425 |
| Erdogan et al. | 37/6/0 | 39/7/2 | 80/6 | 85/11 | 43 | 48 | 0.0513 | 3.797 |
| Maeda et al. | 55/24/0 | 46/15/0 | 134/24 | 107/15 | 79 | 61 | 0.273 | 1.198 |
| This study et al. | 94/22/12 | 100/40/8 | 210/46 | 240/56 | 128 | 148 | 0.147 | 2.097 |
| Wu et al. | 130/76/9 | 93/70/15 | 336/94 | 256/100 | 215 | 178 | 0.723 | 0.125 |
a HWE, Hardy–Weinberg equilibrium.
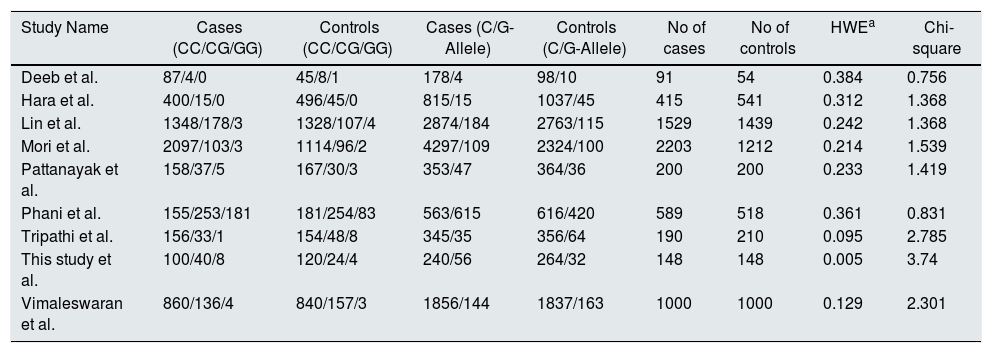
The literature search published from 1991 up to December 2017 retrieved 846 relevant studies from several electronic databases. After removing the duplicate studies, case reports, studies on cell lines, 236 records remained. According to the quality assessment criteria, followed by HWE analysis, nine studies T2DM=6365, controls =5322 participants26–33 were eligible for this meta-analysis. The characteristics (author name, year of publication, country, ethnicity, DNA source, a sample size of cases-controls and genotyping methods) of each study included in the meta-analysis were defined in supplementary Table 2. The genotype, allele frequencies, and HWE/Chi-square values for the studies included in this meta-analysis were presented in Table 7.
Genotype and allele frequencies of PPARγ gene polymorphism with T2DM and controls.
| Study Name | Cases (CC/CG/GG) | Controls (CC/CG/GG) | Cases (C/G-Allele) | Controls (C/G-Allele) | No of cases | No of controls | HWEa | Chi-square |
|---|---|---|---|---|---|---|---|---|
| Deeb et al. | 87/4/0 | 45/8/1 | 178/4 | 98/10 | 91 | 54 | 0.384 | 0.756 |
| Hara et al. | 400/15/0 | 496/45/0 | 815/15 | 1037/45 | 415 | 541 | 0.312 | 1.368 |
| Lin et al. | 1348/178/3 | 1328/107/4 | 2874/184 | 2763/115 | 1529 | 1439 | 0.242 | 1.368 |
| Mori et al. | 2097/103/3 | 1114/96/2 | 4297/109 | 2324/100 | 2203 | 1212 | 0.214 | 1.539 |
| Pattanayak et al. | 158/37/5 | 167/30/3 | 353/47 | 364/36 | 200 | 200 | 0.233 | 1.419 |
| Phani et al. | 155/253/181 | 181/254/83 | 563/615 | 616/420 | 589 | 518 | 0.361 | 0.831 |
| Tripathi et al. | 156/33/1 | 154/48/8 | 345/35 | 356/64 | 190 | 210 | 0.095 | 2.785 |
| This study et al. | 100/40/8 | 120/24/4 | 240/56 | 264/32 | 148 | 148 | 0.005 | 3.74 |
| Vimaleswaran et al. | 860/136/4 | 840/157/3 | 1856/144 | 1837/163 | 1000 | 1000 | 0.129 | 2.301 |
a HWE, Hardy–Weinberg equilibrium.
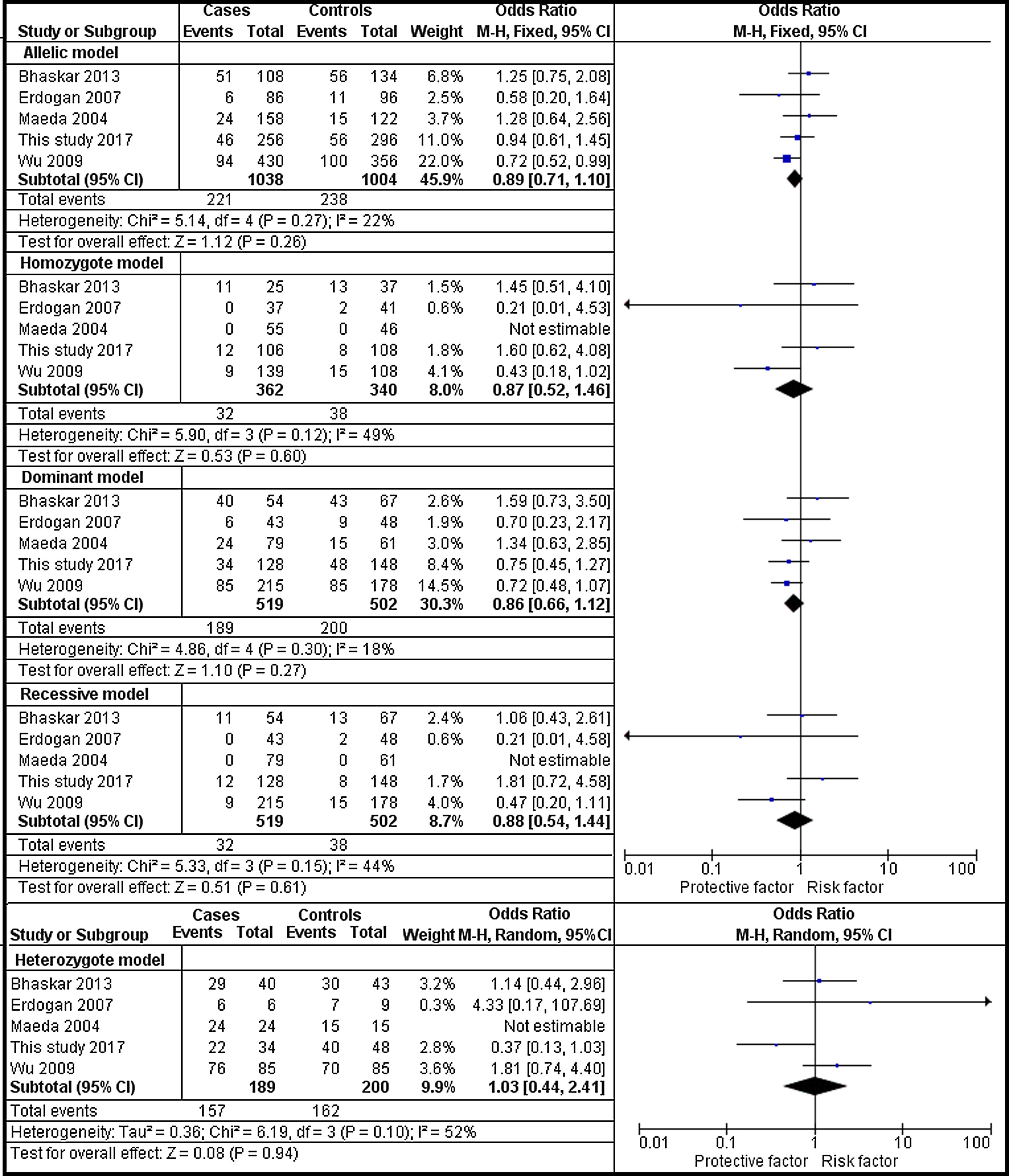
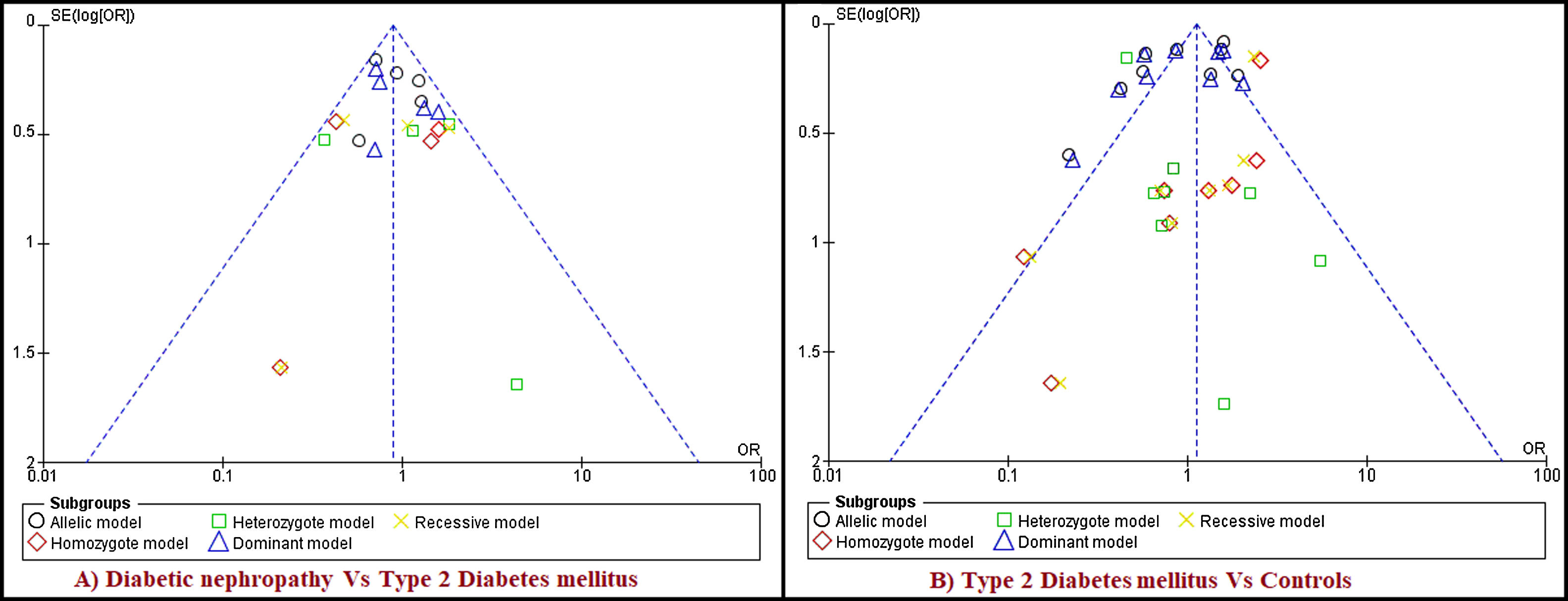
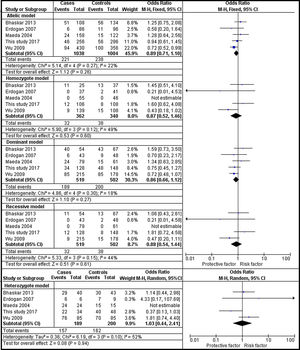
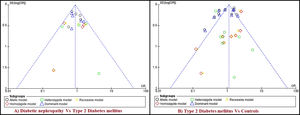
The analysis of rs1801282 polymorphism among DN and T2DM revealed no heterogeneity in allelic (I2=22%) and dominant (I2=18%). Whereas, moderate heterogeneity was noticed in homozygote (I2=49%), heterozygote (I2=52%) and recessive (I2=44%) genetic models. Based on values of heterogeneity (I2) the fixed effects (Mantel–Haenszel's) model was implemented, which showed insignificant association with DN risk in allelic (j vs J) with (Q test p=0.27), OR=0.89, (95%CI [0.71–1.10]), p=0.26, homozygote (jj vs JJ) (Q test p=0.12), OR=0.87, (95% CI [0.66–1.12]), p=0.27, dominant (Jj+jj vs JJ) (Q test p=0.30), OR=0.86, (95% CI [0.66–1.12]), p=0.27 and recessive (jj vs JJ+Jj) (Q test p=0.15), OR=0.88, (95% CI [0.54–1.44]), p=0.61 genetic models, respectively. Whereas, the random effects model was used which showed insignificant association with DN risk in the heterozygote (Jj vs JJ) (Q test p=0.10), OR=1.03, (95% CI [0.44–2.41]), p=0.94. The results of the meta-analysis were shown as forest plots (Fig. 3). Further, funnel plot (Fig. 5) and Egger's test was executed which revealed no publication bias in the studied genetic models. Since a low number of studies were from Caucasian and Asian ethnicity, sub-group meta-analysis was not performed.
Forest plots showing individual and pooled ORs (95% CI) of the risk for rs1801282 polymorphism with Diabetic Nephropathy and Type 2 Diabetes Mellitus. The Forest Plots provides an insignificant association between the PPARγ polymorphism with DN and T2DM under five genetic models. The error bars indicate 95% CIs. Squares represent individual studies in the meta-analysis. Diamonds represent the pooled OR.
Funnel plots of publication biases on the relationship of PPARγ (rs1801282) polymorphism. Funnel plots for publication bias (five genetic models). Each point indicates the individual study included in this meta-analysis. A) Diabetic Nephropathy and Type 2 Diabetes Mellitus B) Type 2 Diabetes Mellitus and Controls.
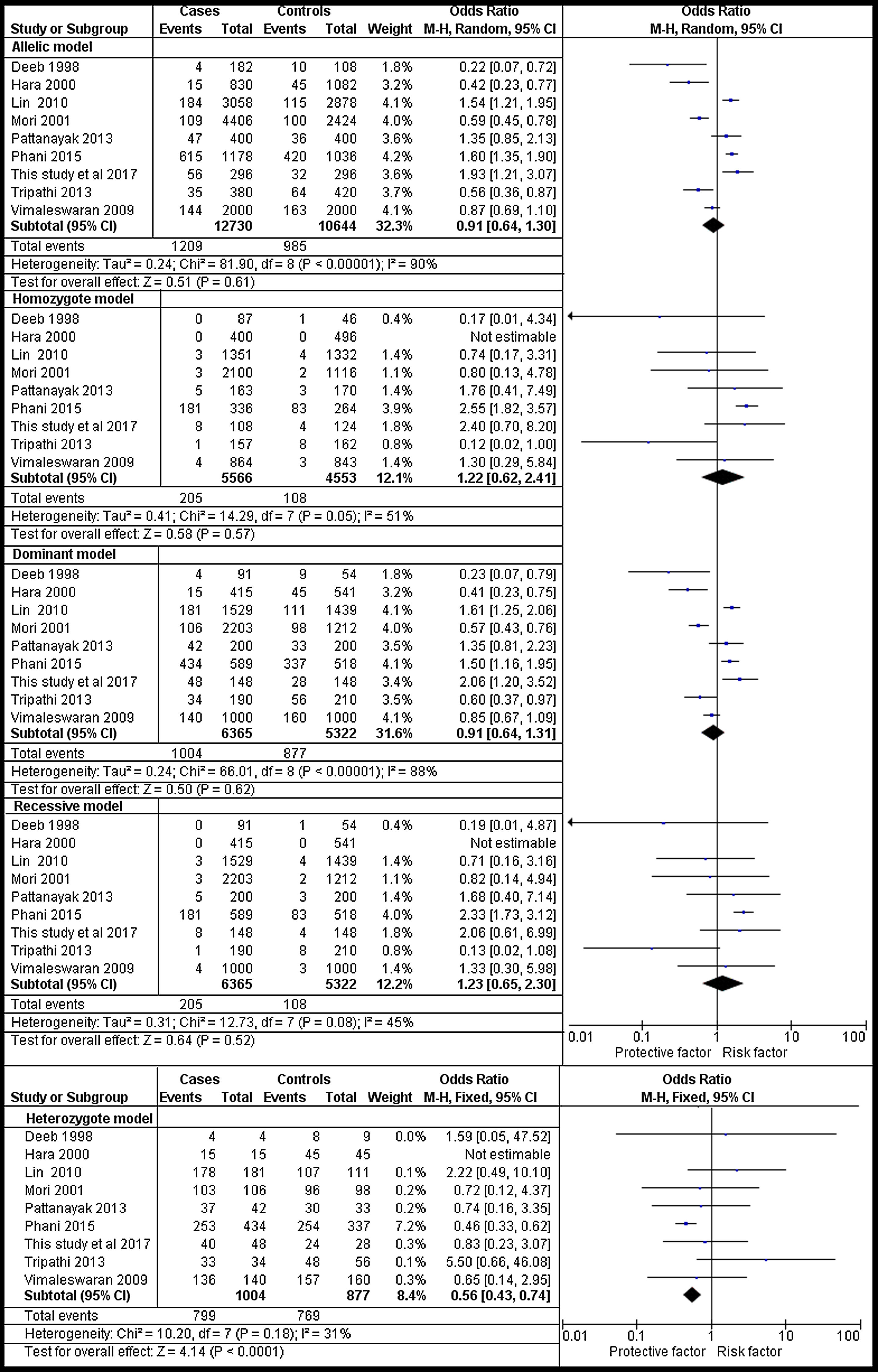
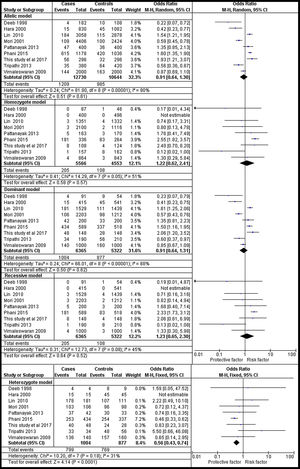
The analysis of rs1801282 polymorphism various T2DM vs. controls revealed moderate heterogeneity in the heterozygote (I2=31%) and high heterogeneity was noticed in allelic (I2=90%), homozygote (I2=51%), dominant (I2=88%) and recessive (I2=45%) genetic models. Based on values of heterogeneity (I2), the random-effect method (DerSimonian and Laird's) was adopted which showed insignificant association with T2DM risk in allelic (j vs J) with (Q test p=0.00001), OR=0.91, (95%CI [0.64–1.30]), p=0.61, homozygote (jj vs JJ) (Q test p=0.05), OR=1.22, (95% CI [0.62–2.41]), p=0.57, dominant (Jj +jj vs JJ) (Q test p=0.00001), OR=0.91, (95% CI [0.64–1.31]), p=0.62 and recessive (jj vs JJ+ Jj) (Q test p=0.08), OR=1.23, (95% CI [0.65–2.30]), p=0.52 genetic models respectively. The fixed effects (Mantel-Haenszel's) model was used, which showed significant association with DN risk in heterozygote (Jj vs JJ) (Q test p=0.18), OR=0.56, (95% CI [0.43–0.74]), p≤0.0001. The results of meta-analysis were illustrated as forest plot (Fig. 4). Further, Begg's funnel plot (Fig. 5) and Egger's regression test were performed which suggested no publication bias in the analyzed genetic models. Since a low number of studies were from Caucasian ethnicity, sub-group analysis was not performed.
Forest plots showing individual and pooled ORs (95% CI) of the risk for rs1801282 polymorphism with Type 2 Diabetes Mellitus and controls. The Forest Plots provides an insignificant association between the PPARγ polymorphism with T2DM and controls under five genetic models. The error bars indicate 95% CIs. Solid squares represent individual studies included in the meta-analysis. Solid diamonds represent the pooled OR.
T2DM is a complex metabolic disorder where environmental, genetic factors and lifestyle contributing to disease pathogenesis.34 The DN susceptibility has been related to various genes localized at different chromosomes. Of various genes, the PPARγ nuclear receptor expressed in adipose tissue and renal glomeruli play a major role in the developmental process of DN.35 The rs1801282 polymorphism has been found to be linked with the reduced ability of trans-activate responsive promoters and thereby lowering the PPAR-γ transcriptional activity.36 On the other hand, PPARs is a potential target for the Glitazones or Thiazolidinediones (TZDs) for the management of T2DM.37
In this study, we have validated the association between PPARγ (rs1801282) polymorphism with the risk of developing DN, T2DM in the south Indian population by comparing 1) DN vs. T2DM, 2) DN vs. controls and 3) T2DM vs. controls. The analysis revealed a positive association with p-value<0.05 between DN and T2DM subjects suggesting that rs1801282 polymorphism might be a risk factor for DN in our study population. These results deviate from a previously published study from the DN subjects of Turkey.23 The analysis of rs1801282 polymorphism among T2DM and controls showed significant association (p-value≤0.05) in derived’G’ allele and heterozygous genotype. These results were in accordance with a previously published study on Japanese subjects belonging to East Asian ethnicity.29
In addition, we have performed a meta-analysis for rs1801282 polymorphism to investigate the effect of the PPARγ gene on susceptibility with DN and T2DM in mixed population. For, which the articles related on DN and T2DM were retrieved from several electronic databases based on PRISMA guidelines and NOS scaling was used for quality assessment. The results of our meta-analysis showed insignificant association with DN in the allelic OR=0.89, (95%CI [0.71–1.10]), homozygote OR=0.87, (95% CI [0.66–1.12]), heterozygote OR=1.03, (95% CI [0.44–2.41]), dominant OR=0.86, (95% CI [0.66–1.12]), and recessive OR=0.88, (95% CI [0.54–1.44]) genetic models respectively. Similar to the results of this meta-analysis, rs1801282 polymorphism shows an insignificant association in Turkey 23 and Taiwan 25 populations.
The meta-analysis of Type 2 Diabetes Mellitus and controls showed significant association among T2DM and controls in the heterozygote genetic model with OR=0.56, (95% CI [0.43–0.74]) and insignificant association allelic OR=0.91, (95%CI [0.64–1.30]), homozygote OR=1.22, (95%CI [0.62–2.41]), dominant OR=0.91, (95%CI [0.64–1.31]), and recessive OR=1.23, (95%CI [0.65–2.30]) genetic models respectively. The results of meta- were in agreement with Japan 29 and Central India 32 whereas disagreement was noticed with Han Chinese 28 population. Overall, the analysis provides significant knowledge on rs1801282 with DN and T2DM in both south Indian and mixed population. However, the results of our analysis can be considered with certain limitations. First, we studied the association between PPARγ genotypes (rs1801282) among DN, T2DM and their phenotypes were not studied extensively. Second, T2DM is a multi-factorial metabolic disorder, other confounding factors including age, gender, obesity, lifestyle habits, and other environmental factors were not considered in our analysis. Third, the studies included in our meta-analysis were restricted to articles published in the English language. Fourth, the genotyping methods were not the same in the included studies. Fifth, we have studied only one SNP (rs1801282) in the PPARγ gene other polymorphic variants were not considered.
ConclusionIn conclusion, our case–control study demonstrates the involvement of PPARγ gene (rs1801282) variant among DN and T2DM in South Indian population. Alternative results were noticed in other population may be due to genetic diversity. However, independent replication studies with larger sample size are required to confirm the associations that we observed in this study. Alternatively, the meta-analysis of DN vs. controls suggests that the PPARγ polymorphism is not a risk factor for developing DN. Whereas, a meta-analysis of T2DM vs. controls suggests that rs1801282 polymorphism result showed a positive association for developing T2DM. Further, well designed, large-scale studies combining with other multiple factors are required to validate the PPARγ gene associated with risk of developing DN and T2DM.
FundingThe author (RI) thanks the Chettinad Academy of Research and Education for funding this research.
Conflicts of interestThe authors declare that they have no conflict of interest.
All the authors were thankful to the patients and controls for participating in the study. The author (AH) wishes to acknowledge the Chettinad Academy of Research and Education (CARE) for providing the Chettinad Research fellowship.