La nefropatía diabética es la causa más común de enfermedad renal crónica terminal. La modulación terapéutica de la angiotensina II retarda, pero no evita, su progresión. La muerte celular contribuye a la pérdida de masa renal en las nefropatías crónicas. Un consorcio europeo empleó la transcriptómica en biopsias renales para identificar nuevos mediadores implicados en la muerte de la célula renal durante la nefropatía diabética. Un 25% de los genes relacionados con la muerte celular estaban expresados diferencialmente en la nefropatía diabética. TRAIL y osteoprotegerina fueron los genes más sobreexpresados, y también estaba aumentado CD74. Las células tubulares y podocitos expresan TRAIL bajo la regulación de citocinas proinflamatorias (MIF vía CD74, TNF). La hiperglucemia sensibiliza a las células renales a la apoptosis inducida por TRAIL, mientras que la osteoprotegerina protege. Estos resultados sugieren que, además de la glucemia, la inflamación y TRAIL pueden ser objetivos terapéuticos en la nefropatía diabética.
Diabetic nephropathy is the most common cause of endstage renal disease. Approaches targeting angiotensin II significantly delay its progression. However, many patients still need renal replacement therapy. High throughput techniques such as unbiased gene expression profiling and proteomics may identify new therapeutic targets. Cell death is thought to contribute to progressive renal cell depletion in chronic nephropathies. A European collaborative effort recently applied renal biopsy transcriptomics to identify novel mediators of renal cell death in diabetic nephropathy. Twenty-five percent of cell death regulatory genes were upor downregulated in diabetic kidneys. TNF-related apoptosisinducing ligand (TRAIL) and osteoprotegerin had the highest level of expression. In diabetic nephropathy, tubular cells and podocytes express TRAIL. Inflammatory cytokines, including MIF via CD74, upregulate TRAIL. A high glucose environment sensitized renal cells to the lethal effect of TRAIL, while osteoprotegerin is protective. These results suggest that, in addition to glucose levels, inflammation and TRAIL are therapeutic targets in diabetic nephropathy.
INTRODUCTION
Chronic kidney disease is characterized by a progressive loss of renal function leading to end-stage renal disease requiring substitution of renal function by dialysis or transplantation. The social and economic costs of these therapies are staggering. In addition, chronic kidney disease shortens survival and many patients die before reaching end-stage renal disease. The pathological substrate of chronic kidney disease is a progressive loss of parenchymal renal cells including glomerular and tubular cells. Apoptosis contributes to renal cell loss and is associated with chronic inflammation and fibrosis.1,2 Angiotensin II is a key mediator of diabetic nephropathy. Preclinical studies indicated that angiotensin II had pleiotropic actions leading to tissue injury and that the renoprotective effects of angiotensin targeting exceeded its anti-hypertensive action.3 A series of randomized controlled trials demonstrated the renoprotective effect of angiotensin converting enzyme inhibitors (ACEI) and angiotensin II receptor antagonists (ARA II) in diabetic nephropathy.4 However, despite these advances, diabetic nephropathy is still the most frequent cause of end-stage renal disease in developed countries. Only a complete understanding of the pathogenic processes that initiate and maintain renal injury will allow the design of novel, successful therapeutic strategies.
HIGH THROUGHPUT TECHNIQUES FOR IDENTIFYING BIOMARKERS AND THERAPEUTIC TARGETS
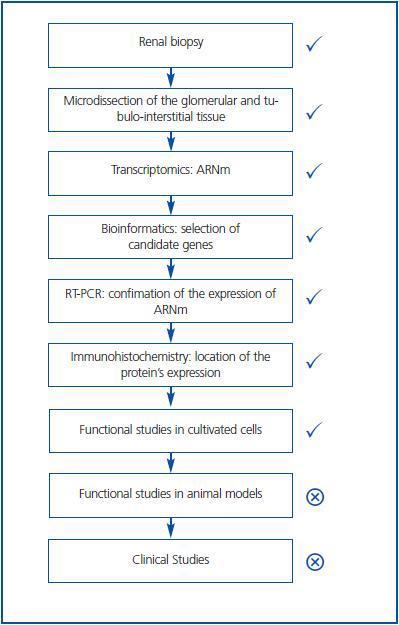
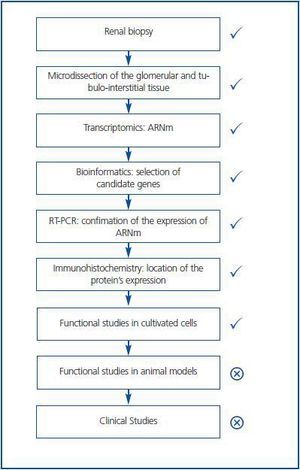
Traditionally the search for mediators of tissue injury entailed the careful scanning of the literature for newly described molecules, which had properties that could make them relevant for renal injury. Once a molecule of potential interest was identified, its expression in renal injury was studied in a one-by-one basis. If protein expression differed between normal and diseased tissue, the actions of the molecule on cultured cells and animal models were explored. This is an inefficient, time-consuming approach. The novel availability of high throughput techniques such as transcriptomics (the simultaneous study of the mRNA expression levels of thousands of genes) or proteomics (the simultaneous study of the expression levels of multiple proteins) allows the identification of hundreds of differentially expressed candidate genes or proteins. Such patterns of expression may themselves be used for diagnostic or prognostic purposes. Bioinformatics algorithms allow to manage the huge amount of data and to search for statistical associations with the presence of disease or with disease progression. Thus, a panel of 65 urine proteomics biomarkers correctly identified diabetic nephropathy with 97% sensitivity and specificity.5 Furthermore, this panel of biomarkers identified patients with microalbuminuria and diabetes that progressed toward overt diabetic nephropathy over 3 years.5 For biomarker identification a pathogenic function of the candidate is not required. In addition, novel therapeutic targets may be uncovered. For this, bioinformatics and focused data analysis help to prioritize molecules with highest or lowest level of expression, to identify molecules related to processes that participate in the pathogenesis of the disease, and to identify relationships between differentially regulated molecules. This reduces the field of candidate therapeutic targets. Then, the differential expression of these candidates can be confirmed by RT-PCR at the mRNA level and by immunohistochemistry at the protein level (Figure 1.) Immunohistochemistry localizes the cell types expressing the protein, thus guiding the choice of cell culture models. Additional criteria to choose a molecule for further studies may be the simultaneous differential expression of several molecules that belong to the same functional pathway. This was the case for cell death related genes of the TNF-related apoptosis-inducing ligand (TRAIL) pathway expressed in human diabetic nephropathy.6
TRANSCRIPTOMICS OF DIABETIC KIDNEYS
The European Renal cDNA Bank (ERCB) was initially funded by the European Union and is now located at the University of Zurich (Nephrology Clinic and Institute of Physiology) (http://www.portailderecherche.ch/unizh/p9291.htm.) Fragments of renal biopsies were collected from all over Europe and classified by pathological diagnosis. Tubulointerstitial and glomerular compartments were manually microdissected from cortical tissue segments and mRNA expression was assessed by microarray chips displaying 22,283 probe sets. The resulting huge amount of gene expression data was used to further unravel the pathogenesis of diabetic kidney injury. As an example, the activation of the NFkB pathway was tested in human diabetic nephropathy. The focused analysis of this master transcriptional switch, involved in multiple processes, segregated progressive diabetic nephropathy from mild diabetic nephropathy and controls by showing upregulation of 54 of 138 known NF-kB targets.7
APOPTOSIS-RELATED GENE EXPRESSION IN HUMAN DIABETIC NEPHROPATHY
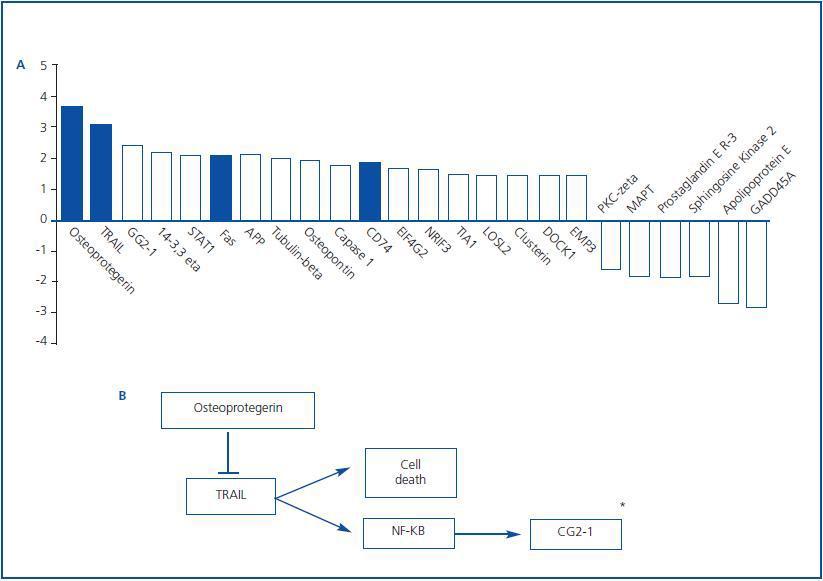
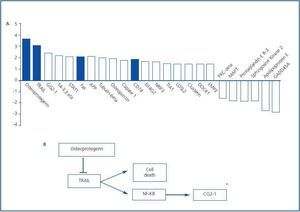
Apoptotic cell death may contribute to the gradual loss of renal mass in diabetic nephropathy.1,8 Indeed, 25% of apoptosis related genes were differentially regulated in the tubulointerstitium of renal biopsies from diabetic nephropathy patients6. Those with a higher than 1.5 fold change are presented in figure 2. Differentially expressed apoptosisrelated genes included those encoding proteins involved in death receptor interactions, such as Fas, TRAIL and osteoprotegerin (OPG.) These data are consistent with the already described upregulation of Fas in diabetic nephropathy.9 Cytokines belonging to the TNF superfamily are key regulators of cell survival, inflammation and fibrosis. Among them, Fas ligand and TWEAK participate in the pathogenesis of renal injury and TNF has an important role in diabetic nephropathy.10-15 TNF may promote synthesis and release of chemokines and inflammatory and hemodynamic mediators, proliferation, cytotoxicity and regulation of cell death related genes.15 However, the role of TRAIL and OPG in kidney disease was unexplored.
TRAIL
TRAIL (APO2L/TNFSF10) is a TNF superfamily cytokine that binds to a complex system of receptors.16-18 Depending on the relative levels of the TRAIL receptors and on the cellular system, TRAIL can exert different functions that, in addition to cell death, also include survival, proliferation, and maturation.19 TRAIL is a type II trans-membrane protein with a molecular mass of 33-35 kDa. Membrane-bound TRAIL can be cleaved from the cell surface to form a soluble trimeric ligand that retains the proapoptotic activity. TRAIL induces apoptosis in human cancer cells and primary tumors, showing minimal or absent toxicity on normal cells. TRAIL and TRAIL-receptor (TRAIL-R) agonists are in clinical trials as anti-cancer agents.20 TRAIL is normally expressed in many human tissues including liver, colon, heart, kidney, lung and testis, which is consistent to the low cytotoxicity to most healthy tissues.21 However, data from knockout mice suggest that TRAIL may induce apoptosis in non-tumoral parenchymal cells immersed in an inflammatory environment.22 Available studies on TRAIL and diabetes have emphasized its role in immune response regulation.23
TRAIL RECEPTORS
TRAIL has a complex system of receptors. In humans, it consists in 4 membrane-bound and a soluble receptor. All TRAIL receptors belong to the TNF receptor superfamily. Mice have only one receptor for TRAIL with a functional DD, but the system is very similar24 Human TRAIL-R1 (DR4/TNFRSF10A) and TRAIL-R2 (DR5/TRICK2/KILLER/TNFSFR10B) contain a cytoplasmic death domain and activate caspases, promoting apoptosis.18 By contrast, human TRAIL-R3 (TRID/DcR1/ LIT/TNFRSF10C) and TRAIL-R4 (DcR2/TRUNDDTNFRSF10D) bind TRAIL without activation of the apoptotic machinery and are considered decoy receptors.25 TRAIL-R3 has a glycosylphosphatidylinositol membrane anchor and lacks an intracellular domain and TRAIL-R4 contains a truncated DD. OPG (TNFRSF11B) is a soluble decoy receptor for TRAIL.26 In addition OPG binds to and antagonizes the TNF superfamily member receptor activator of NF-kB ligand (RANKL).27 As OPG is a decoy for both TRAIL and RANKL potential cross-regulatory mechanisms involving the balance among TRAIL, OPG and RANKL may exist. Indeed, OPG binding to TRAIL inhibits its interaction with TRAIL-Rs and consequently TRAIL-induced apoptosis. Conversely, TRAIL can block the inhibitory activity of OPG on osteoclastogenesis.17 The affinity of TRAIL for OPG is weaker than that for its lethal receptors, but recent studies support the biological relevance of the OPG/TRAIL interaction.20 OPG is increased in serum of patients with renal dysfunction, including diabetic nephropathy.28-30 However, the source of serum OPG is unclear. Increased serum OPG levels are also associated to increased coronary artery and aorta calcification.30 OPG deficient mice display calcification of the aorta and renal arteries, suggesting that OPG plays a role in vascular calcification.26
TRAIL AND ITS RECEPTORS IN THE KIDNEY
TRAIL, TRAIL-R1 and -R2 mRNA are expressed in the normal kidney. TRAIL is expressed in tubules but not in normal glomeruli. TRAIL-R1 has a similar pattern of expression, while TRAIL-R2 is additionally expressed in Henle´s loop. TRAIL-R3 and R4 are absent from or not studied in normal kidney.31 TRAIL knock-out mice did not have renal pathology, suggesting that TRAIL is not essential for normal kidney physiology and development.
INTERPLAY OF HIGH GLUCOSE LEVELS AND INFLAMMATION IN DIABETIC KIDNEY INJURY
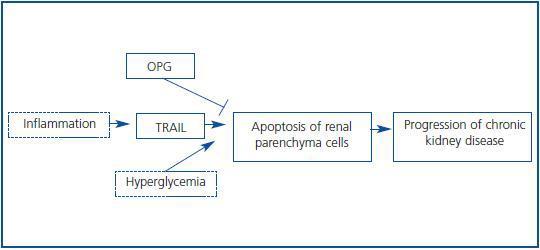
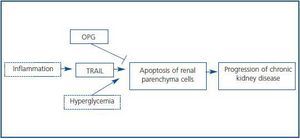
The transcriptomic finding of increased expression of TRAIL and OPG in human diabetic kidneys was confirmed by RT-PCR and found to correlate to clinical severity of renal disease.6 In an independent cohort of diabetic nephropathy patients TRAIL protein was increased as assessed by immunohistochemistry. In diabetes tubular epithelium was the main site of TRAIL expression, and there also was de novo podocyte expression.6,32 Furthermore TRAIL staining correlated with tubular atrophy, interstitial fibrosis and interstitial inflammation, suggesting a pathogenic role for TRAIL in diabetic kidney injury. Both TRAIL and OPG, two potentially antagonic molecules, were upregulated, and their relationship was dissected in cultured human tubular cells (Figure 3.) First, factors contributing to increased TRAIL expression were studied. High glucose per se did not modulate TRAIL expression. However, proinflammatory cytokines present in chronic kidney injury, such as IFNg and TNFa,15,33 increased TRAIL expression. CD74 is a receptor for MIF overexpressed in human diabetic nephropathy (Figure 2.) Its engagement by MIF increased TRAIL expression in cultured human podocytes and tubular cells.32 Next we approached possible functions of TRAIL in tubular cells. TRAIL weakly induced tubular cell death in a dose-dependent manner6. This is consistent with a role in a nephropathy that progresses over years, such as diabetic nephropathy. The environment modulated the sensitivity of tubular cells to TRAIL-induced cell death. The combination of high glucose and proinflammatory cytokines further increased the susceptibility of tubular cells to TRAIL-induced apoptosis. Hyperglycemia is among the microenvironmental factors that may induce or facilitate apoptosis.8,34,35 Several factors contribute to the higher sensitivity of tubular cells in a diabetic or inflammatory milieu to apoptosis. High expression of death receptors (Fas, Fn14) and intracellular lethal molecules (Bax, Smac/Diablo, FADD), as well as decreased expression of antiapoptotic molecules (Bcl2, BclxL) have been observed.11,14,36-38 We are currently characterizing the role of additional molecules, not previously suspected to be involved in cell death, that have been identified by transcriptomics and functional genomics.32,39
NFkB activation by TRAIL also protected cells from lethality induced by TRAIL.6 In this regard, TRAIL resembles TNF in that it activates simultaneously death and survival signals. If NFkB-mediated survival signals are blocked, cell death increases. Finally OPG protected from TRAIL-induced cell death by behaving as a decoy receptor. We hypothesize that the end result of the increased expression of TRAIL and OPG in a particular patient may depend on the relative balance between them (Figure 3.) If TRAIL prevails, tissue injury results. If OPG prevails, the cells are protected from TRAIL. Nevertheless, OPG may have additional functions in diabetic nephropathy.
The expression of TRAIL, TRAIL-R1 and -R2 increases in renal proximal and distal convoluted tubules of rejected kidney tissue.40 However, there is insufficient information on the expression of TRAIL and its receptors in other inflammatory renal diseases.
FOLLOWING THE TRAIL
In medicine every answer brings new questions. TRAIL is the most upregulated gene encoding proapoptotic proteins in human diabetic kidneys. We now have to explore what additional factors regulate its expression in tubular cells and podocytes, how does glucose sensitize to the proapoptotic effect of TRAIL and whether TRAIL therapeutic modulation protects from the development or progression of diabetic nephropathy in preclinical studies. In addition, the biomarker potential of molecules in this pathway should be explored.
Figure 1.
Figure 2.
Figure 3.