The introduction of donor-derived free DNA (ddcfDNA) has emerged as an accurate non-invasive biomarker to diagnose rejection, compared to classical ones. Here we evaluate our experience after its implementation in our center as an in-house technique.
Materials and methodsSingle-center cross-sectional study with extraction of cell-free DNA in blood and quantification of the ddcfDNA using the AlloSeqcfDNA assay (CareDx) at the time of performing biopsies ‘per protocol’ or ‘per indication’ between December 2020 and December 2023.
Results172 graft biopsies were included (59 for protocol and 113 for cause) in 112 kidney transplant recipients. Among the biopsies, 19 borderline rejections, 11 T-cell mediated rejections, and 30 antibody-mediated rejections were identified. The median ddcfDNA in each diagnostic group was: 0.40% (0.23%–0.82%) in borderline, 0.60% (0.23%–1.91%) in cellular, and 1.48% (0.77%–3.4%) in antibody-mediated rejection (P < .001). In the 112 biopsies with no signs of rejection, the median ddcfDNA was 0.33% (0.17%–0.54%) (P < .001). Cases with positive DSAs and rejection showed higher levels of ddcfDNA than positive DSAs without rejection (P = .010), and ddcfDNA levels were significantly associated with microvascular inflammation and C4d positivity. The area under the ROC curves of ddcfDNA to discriminate any type of rejection from the absence of rejection was 0.74 (0.65−0.82) and, excluding borderline rejection from the analysis, 0.80 (0.72−0.89), outperforming other markers of renal function.
ConclusionsImplementing ddcfDNA analysis at our center as a clinical tool has proven valuable for distinguishing biopsy-confirmed acute rejection, particularly antibody-mediated rejection, outperforming classic renal function markers. Its hospital-based implementation supports timely and accurate diagnosis, improving transplant management and prognosis.
El único método actual definitivo para el diagnóstico de rechazo en el paciente trasplantado renal es mediante la biopsia del injerto. La introducción del ADN libre derivado del donante (ddcfDNA) ha emergido como un biomarcador no invasivo más preciso para cuantificar esta lesión y diagnosticar el rechazo, en comparación con biomarcadores clásicos. Aquí evaluamos nuestra experiencia tras su implementación en nuestro centro.
Materiales y métodosEstudio transversal unicéntrico con extracción de cell-free DNA en sangre y cuantificación mediante ensayo AlloSeqcfDNA (CareDx) del ddcfDNA en el momento de realización de biopsias «por protocolo» o «por indicación» entre diciembre 2020 y diciembre 2023.
ResultadosEn total se incluyeron 172 biopsias de injerto (59 por protocolo y 113 por indicación) en 112 receptores de trasplante renal. Entre las biopsias, se identificaron 19 rechazos borderline, 11 rechazos mediados por células T y 30 rechazos mediados por anticuerpos. La mediana de ddcfDNA en cada grupo diagnóstico fue: 0,40% (0,23%–0,82%) en el borderline, 0,60% (0,23%–1,91%) en el celular y 1,48% (0,77%–3,4%) en el mediado por anticuerpos (P < ,001). En las 112 biopsias sin datos de rechazo, la mediana de ddcfDNA fue de 0,33% (0,17%–0,54%) (P < ,001). Los casos con DSAs positivos y rechazo presentaron niveles de ddcfDNA más elevados en comparación con los DSAs positivos sin rechazo (P = ,010) y los niveles de ddcfDNA mostraron una asociación significativa con la inflamación microvascular y la positividad de C4d. El área bajo las curvas ROC del ddcfDNA para discriminar cualquier tipo de rechazo de la ausencia de rechazo fue de 0,74 (0,65–0,82) y excluyendo el rechazo borderline del análisis, de 0.80 (0.72–0.89), superando a otros marcadores de función renal.
ConclusionesLa implementación del análisis ddcfDNA en nuestro centro como herramienta asistencial ha demostrado ser valiosa para distinguir el rechazo agudo comprobado por biopsia, especialmente el mediado por anticuerpos, superando a otros marcadores clásicos de función renal. Su implementación hospitalaria favorece un diagnóstico oportuno y preciso, mejorando la gestión y pronóstico del trasplante.
Antibody-mediated rejection (ABMR) remains the most relevant challenge to ensure long-term renal graft survival. Early diagnosis is essential, as treatments are ineffective when acute rejection progresses to a chronic form.1 Unfortunately, biomarkers such as creatinine and proteinuria respond late, once rejection is established, and donor-specific antibodies (DSA) are not highly sensitive, as almost half of ABMR cases occur without detectable DSA.2
A recently introduced biomarker that could transform clinical practice is donor-derived cell-free DNA (ddcfDNA). Donor cells release ddcfDNA by apoptosis or necrosis and ddcfDNA constitutes a percentage of the recipient's circulating cell-free DNA. When graft damage occurs, that percentage increases significantly, making it a useful, sensitive marker of damage.3 However, it is not a specific biomarker of rejection, but of graft cell damage, since elevated levels occur in other circumstances such as acute tubular necrosis (ATN)4 and BK virus infection.5
Several studies have highlighted the diagnostic capacity of ddcfDNA.1,6–9 Several clinical analysis service providers perform the test by sending samples to a centralized laboratory or in-house using massive next-generation sequencing (NGS) techniques. Hospital Clínic of Barcelona implemented in-house ddcfDNA detection at the end of 2020.
The objective of this study is to analyze the center's experience in the implementation of this technique, as well as the clinical results obtained since.
Material and methodsStudy design and populationThis is a single-center cross-sectional study at the Hospital Clínic of Barcelona, in which we analyzed blood and histological samples from renal transplant patients who underwent a graft biopsy between December 2020 and December 2023 and determined in parallel the percentage of ddcfDNA in blood. The analysis included biopsies performed both by clinical indication, due to abnormalities in analytical results (creatinine alterations, decrease in estimated glomerular filtration rate [eGFR] by CKD-EPI equation or worsening proteinuria), accompanied or not by the development of de novo DSA, and biopsies performed by protocol. In our center, these are routinely performed at 3 and 12 months post-transplantation, regardless of graft function. We collected and analyzed demographic, clinical, analytical, and immunological data of the included patients. We sent all biopsies for evaluation by the Anatomic Pathology Service of the Hospital Clínic de Barcelona, following the Banff 2019 classification. To ensure the internal validity of the results, we excluded patients who had received multiple organ transplants, avoiding possible interferences derived from other grafts.
Quantification of donor-derived cell-free DNA (ddcfDNA) by massive sequencingIn each patient, blood was collected in 2 10 ml Streck tubes (Streck, La Visa, NE, USA), which were kept at room temperature and centrifuged within 7 days after sample collection. Plasma obtained from centrifugation (the first at 1600 g for 20 min, and the second at 16,000 g for 10 min) was stored at –80 °C until further analysis. Cell-free DNA was extracted from 5 ml of plasma using the QiAmp circulating nucleic acid kit (Qiagen, Dusseldorff, Germany) following the manufacturer's instructions. The relative amount of cell-free DNA from the donor in the kidney transplant recipient was assessed by mass sequencing of 202 single nucleotide polymorphisms (SNPs). Libraries were prepared using the AlloSeq cfDNA Assay (CareDx, San Francisco, CA, USA) following the manufacturer's protocol and sequencing was performed on a MiSeq instrument using the 300-cycle MiSeq Micro v2 or 150-cycle MiSeq v3 kits (Illumina, San Diego, CA, USA) depending on the number of samples to be processed. Data analysis was performed with AlloSeq cfDNA v2.2.1 software (CareDx) using blind analysis which does not require genotyping of any genomic samples, and ddcfDNA was measured as the percentage of all available cfDNA in the sample.
Statistical analysisThe normality of continuous variables was assessed using the Kolmogorov-Smirnov test. Data for quantitative variables with normal distribution are presented as mean and standard deviation (SD), while for those with non-normal distribution we used Turkey’s method to calculate the median and interquartile range (IQR). Qualitative variables are described with absolute and relative frequencies.
Depending on the nature of the data, Student's t-test was used to compare variables with normal distribution, while for variables with non-normal distribution, Mann-Whitney U or Kruskal-Wallis tests were applied. In the analysis of categorical data, Chi-square or Fisher tests were applied, as appropriate. For multivariate analysis, binary logistic regression was performed.
The area under the ROC curves (AUC) was used to evaluate the performance of ddcfDNA, DSA, eGFR and urine protein/creatinine ratio (UPCR) in the diagnosis of rejection. The AUC results include their 95% confidence interval.
Statistical analysis was performed with IBM SPSS Statistics version 21.0 software (SPSS, Inc; Chicago, Illinois) for Windows, and the graphical representations were designed with GraphPad version 9.5.1 (GraphPad Software, La Jolla, CA, USA). Two-tailed statistical significance tests were applied and a significance levels of P < .05 was established.
ResultsThis study included a total of 172 renal biopsies, performed on 112 renal transplant recipients, of which 59 (34.3%) were protocol biopsies and 113 (65.7%) were clinically indicated biopsies performed at 24 (4–68) weeks post-transplantation.
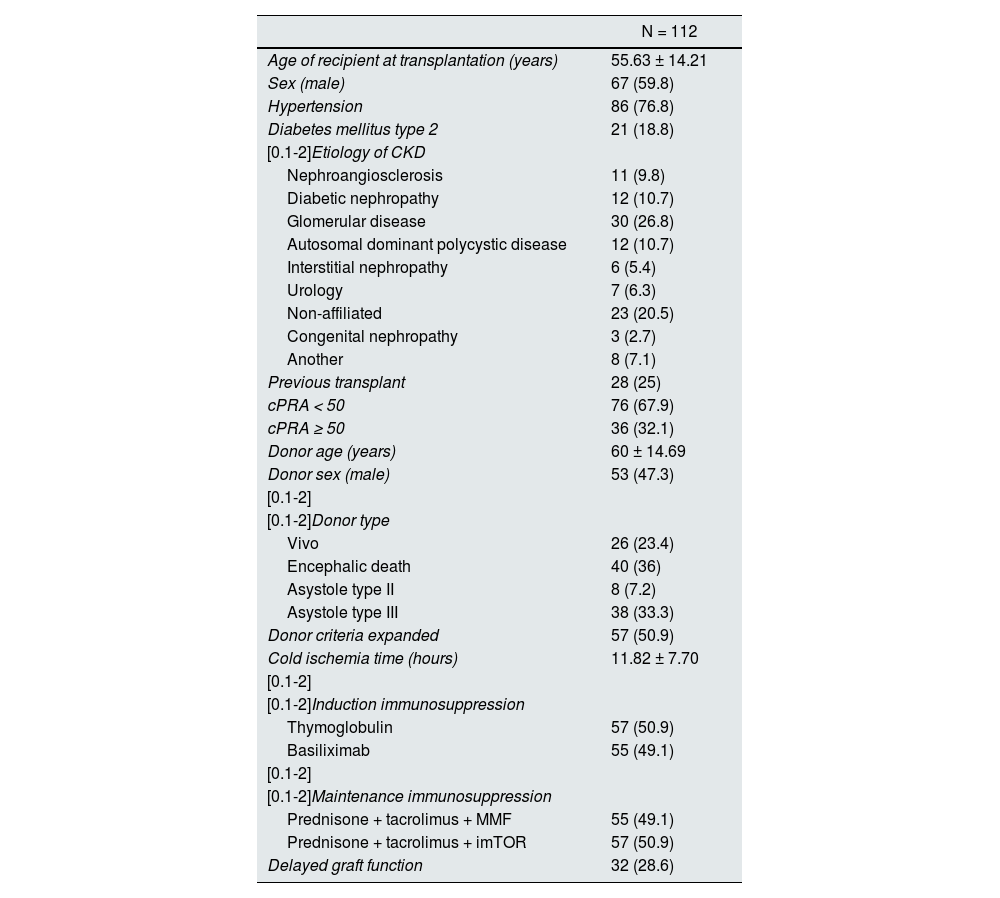
Among the 112 recipients, 26 (23.4%) received a graft from a living donor, 40 (36%) from a brain-dead donor, 38 (33.3%) from a type III asystole donor and 8 (7.2%) from a type II asystole donor. The mean age was 58.78 ± 13.45 years and 59.8% of patients were male. Among previous transplants, 87 patients (77.7%) were first-time transplant recipients, 13 (11.6%) second-time, 6 (5.4%) third-time, 4 (3.6%) fourth-time and 2 patients (1.8%) were fifth-time transplant recipients. The main baseline characteristics of the patients are in Table 1.
Main baseline characteristics of the included patients.
| N = 112 | |
|---|---|
| Age of recipient at transplantation (years) | 55.63 ± 14.21 |
| Sex (male) | 67 (59.8) |
| Hypertension | 86 (76.8) |
| Diabetes mellitus type 2 | 21 (18.8) |
| [0.1-2]Etiology of CKD | |
| Nephroangiosclerosis | 11 (9.8) |
| Diabetic nephropathy | 12 (10.7) |
| Glomerular disease | 30 (26.8) |
| Autosomal dominant polycystic disease | 12 (10.7) |
| Interstitial nephropathy | 6 (5.4) |
| Urology | 7 (6.3) |
| Non-affiliated | 23 (20.5) |
| Congenital nephropathy | 3 (2.7) |
| Another | 8 (7.1) |
| Previous transplant | 28 (25) |
| cPRA < 50 | 76 (67.9) |
| cPRA ≥ 50 | 36 (32.1) |
| Donor age (years) | 60 ± 14.69 |
| Donor sex (male) | 53 (47.3) |
| [0.1-2] | |
| [0.1-2]Donor type | |
| Vivo | 26 (23.4) |
| Encephalic death | 40 (36) |
| Asystole type II | 8 (7.2) |
| Asystole type III | 38 (33.3) |
| Donor criteria expanded | 57 (50.9) |
| Cold ischemia time (hours) | 11.82 ± 7.70 |
| [0.1-2] | |
| [0.1-2]Induction immunosuppression | |
| Thymoglobulin | 57 (50.9) |
| Basiliximab | 55 (49.1) |
| [0.1-2] | |
| [0.1-2]Maintenance immunosuppression | |
| Prednisone + tacrolimus + MMF | 55 (49.1) |
| Prednisone + tacrolimus + imTOR | 57 (50.9) |
| Delayed graft function | 32 (28.6) |
Data are expressed as mean ± standard deviation or absolute number and percentage.
cPRA: percentage of reactivity against calculated panel; imTOR: mTOR inhibitors; MMF: mycophenolate mofetil.
Regarding histopathological findings, we identified rejection in 60 (34.9%) of the 172 biopsies, the majority for clinical indication (52, 86.7%) rather than protocol biopsies (8, 13.3%), P < .001. Among the 112 biopsies without rejection, 61 (54.5%) were requested by clinical indication and 51 (45.5%) by protocol.
Of the 60 cases with rejection, 19 (31.67%) were borderline rejection cases, 11 (18.33%) T cell mediated rejection (TCMR) cases and 30 (50%) were ABMR cases. Regarding TCMR classification, 4 cases were type IA rejection, 2 type IB, 2 type IIA and 3 chronic-active rejection. In the case of ABMR, 21 were acute rejections and 9 chronic-active rejections. We detected DSA in 19 of the 30 ABMR cases (63.3%).
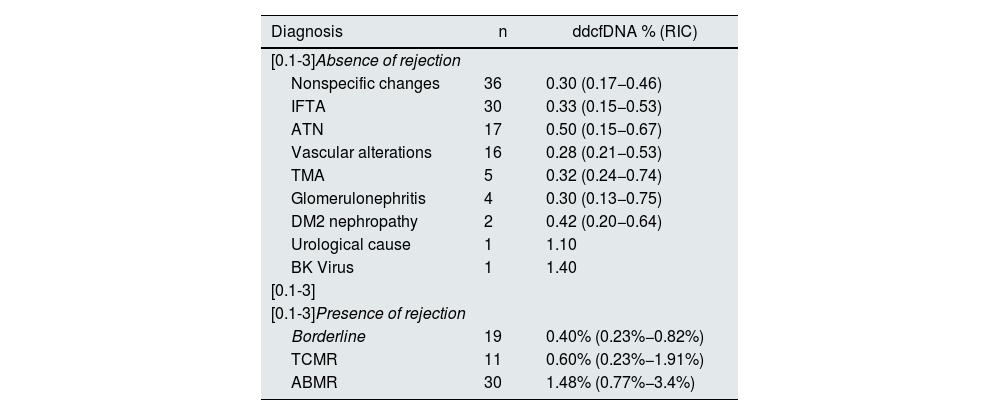
Among the biopsies without rejection, the most common diagnoses were nonspecific changes (36 cases, 20.9%), interstitial fibrosis and mild to moderate tubular atrophy (30 cases, 17.4%), ATN (17 cases, 9.9%) and vascular alterations (16 cases, 14.3%). Other diagnoses included thrombotic microangiopathy (5 cases, 2.9%), glomerulonephritis (4 cases, 2.3%), complications related to type 2 diabetes (2 cases, 1.2%), urological cause (1 case, 0.6%) and BK virus infection (1 case, 0.6%). Table 2 shows the median ddcDNA in each diagnosis.
ddcDNA values in cases of absence or presence of rejection.
| Diagnosis | n | ddcfDNA % (RIC) |
|---|---|---|
| [0.1-3]Absence of rejection | ||
| Nonspecific changes | 36 | 0.30 (0.17−0.46) |
| IFTA | 30 | 0.33 (0.15−0.53) |
| ATN | 17 | 0.50 (0.15−0.67) |
| Vascular alterations | 16 | 0.28 (0.21−0.53) |
| TMA | 5 | 0.32 (0.24−0.74) |
| Glomerulonephritis | 4 | 0.30 (0.13−0.75) |
| DM2 nephropathy | 2 | 0.42 (0.20−0.64) |
| Urological cause | 1 | 1.10 |
| BK Virus | 1 | 1.40 |
| [0.1-3] | ||
| [0.1-3]Presence of rejection | ||
| Borderline | 19 | 0.40% (0.23%−0.82%) |
| TCMR | 11 | 0.60% (0.23%−1.91%) |
| ABMR | 30 | 1.48% (0.77%−3.4%) |
Data are expressed as median (IQR: interquartile range).
ABMR, antibody-mediated rejection; ddcfDNA, donor-derived cell-free DNA; DM2, type 2 diabetes mellitus; IFTA, interstitial fibrosis and tubular atrophy; TMA, thrombotic microangiopathy; ATN, acute tubular necrosis; TCMR, T-cell-mediated rejection.
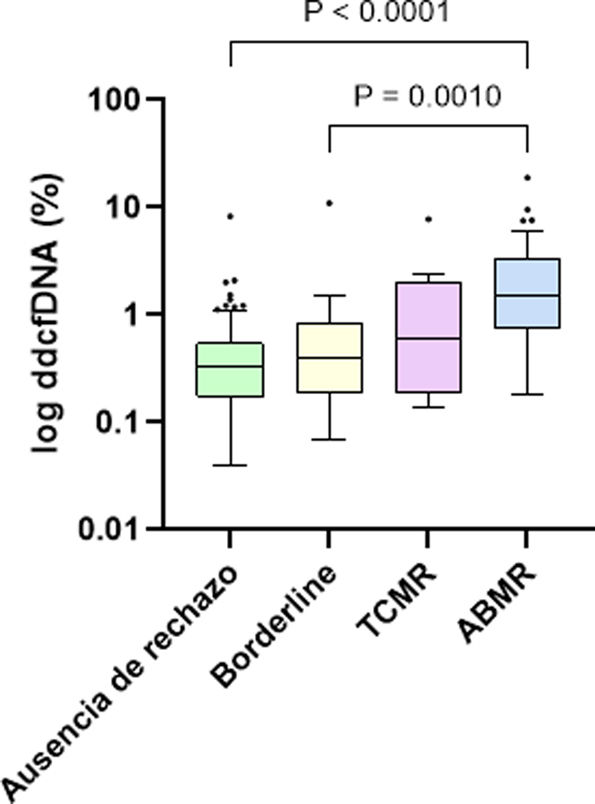
Regarding ddcfDNA levels, a significant difference was observed between patients with and without rejection, with a median of 0.88% (0.34%–2.08%) in rejection cases and 0.33% (0.17%–0.54%) in the absence of rejection (P < .001). Specifically, the median ddcfDNA was 0.40% (0.23%–0.82%) in borderline cases, 0.60% (0.23%–1.91%) in TCMR and 1.48% (0.77%–3.4%) in ABMR, evidencing significantly higher values in patients with ABMR compared to patients without rejection (P < .001) and patients with borderline rejection (P = .0010) (Fig. 1). Within TCMR, there was no significant difference in ddcfDNA levels between IA and ≥IB rejections (P = .788). There were also no significant differences between acute and chronic-active ABMRs (1.59% vs. 0.97%, P = .108).
The cases with positive DSA and rejection data at biopsy (19 ABMR and 2 TCMR) showed significantly higher levels of ddcfDNA compared to the cases with positive DSA but no rejection (n = 8), with a median of 2.13% (1.2%–4.8%) vs. 0.69% (0.28%–1.23%) (P = .010). In addition, the 19 cases with positive DSA and ABMR at biopsy also had significantly higher median ddcfDNA (2.34%; 1.28%–5.47%) compared to the 11 ABMR cases without DSA (0.97%; 0.52%–1.24%), with a P-value of .010.
Regarding eGFR and UPCR, no significant differences were observed between patients with and without rejection (30.31 ± 16.98 mL/min/1.73 m2 vs. 36.18 ± 19.64 mL/min/1.73 m2, P = .135; 522 mg/g vs. 359 mg/g, P = .171, respectively).
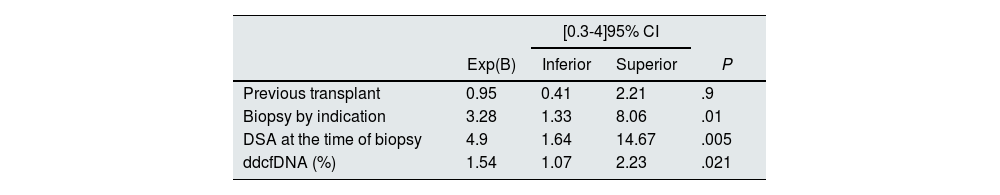
In univariate analysis, the variables significantly associated with rejection were indication biopsy (P < .001), presence of DSA (P < .001), previous transplant (P = .023) and ddcfDNA levels (P < .001). In multivariate analysis, indication biopsy (P = .010), DSA at the time of biopsy (P = .005) and ddcfDNA levels (P = .021) maintained their association, while previous transplantation (P = .9) lost its association with rejection (Table 3).
Multivariate analysis of the variables associated with rejection.
| [0.3-4]95% CI | ||||
|---|---|---|---|---|
| Exp(B) | Inferior | Superior | P | |
| Previous transplant | 0.95 | 0.41 | 2.21 | .9 |
| Biopsy by indication | 3.28 | 1.33 | 8.06 | .01 |
| DSA at the time of biopsy | 4.9 | 1.64 | 14.67 | .005 |
| ddcfDNA (%) | 1.54 | 1.07 | 2.23 | .021 |
ddcfDNA: donor-derived cell-free DNA; DSA: donor-specific antibodies.
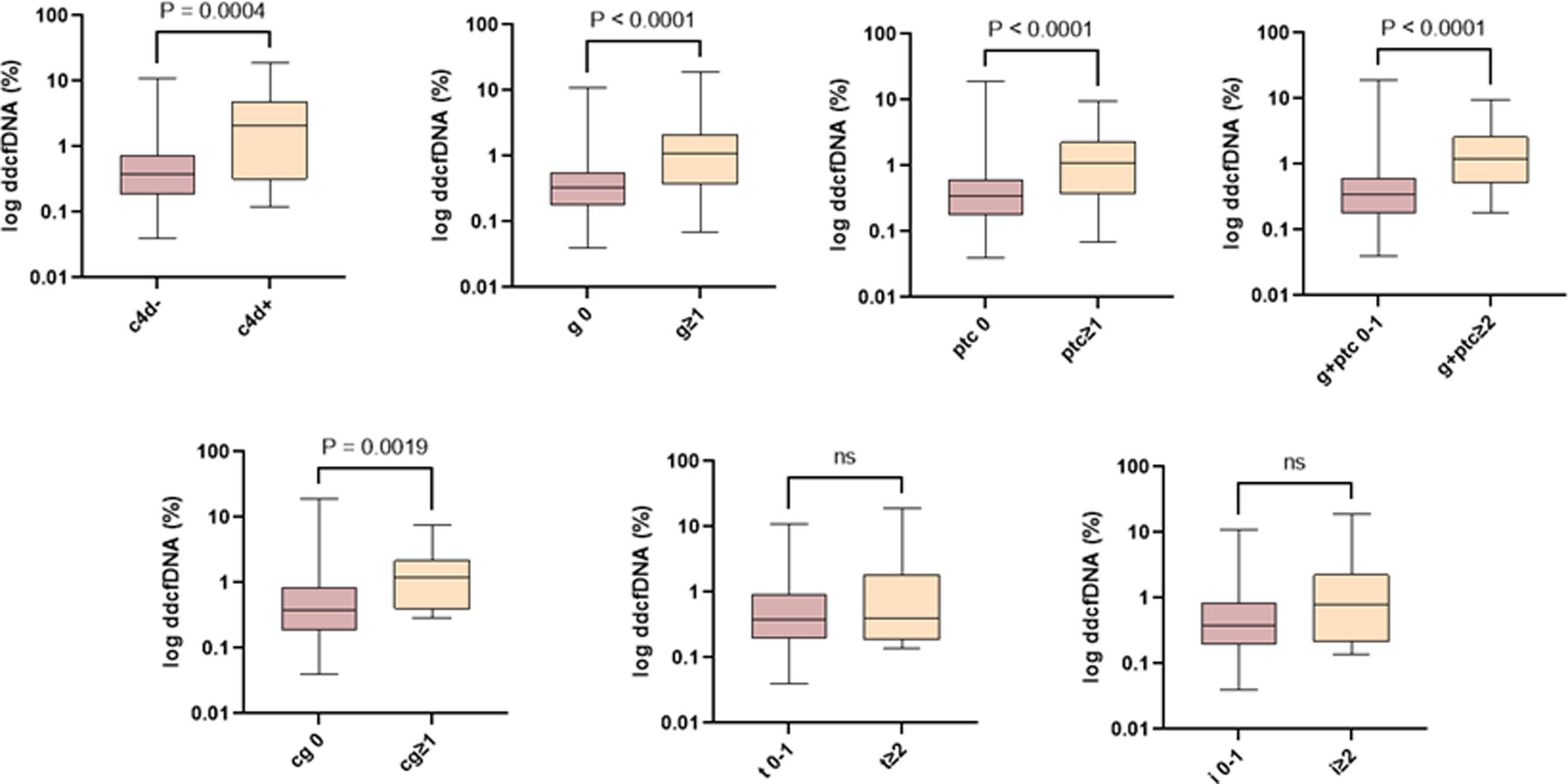
In the association analysis of ddcfDNA with the individual scores of the Banff classification, we observed a significant correlation with microvascular inflammation and the presence of positive C4d, but not with tubulointerstitial inflammation (Fig. 2).
Association of ddcfDNA levels with individual Banff classification scores. The ddcfDNA correlates with the severity of individual lesions associated with an alloimmune lesion. ddcfDNA: donor-derived cell-free DNA; cg: transplant glomerulopathy; g: glomerulitis; i: interstitial inflammation; ptc: peritubular capillaritis; t: tubulitis.
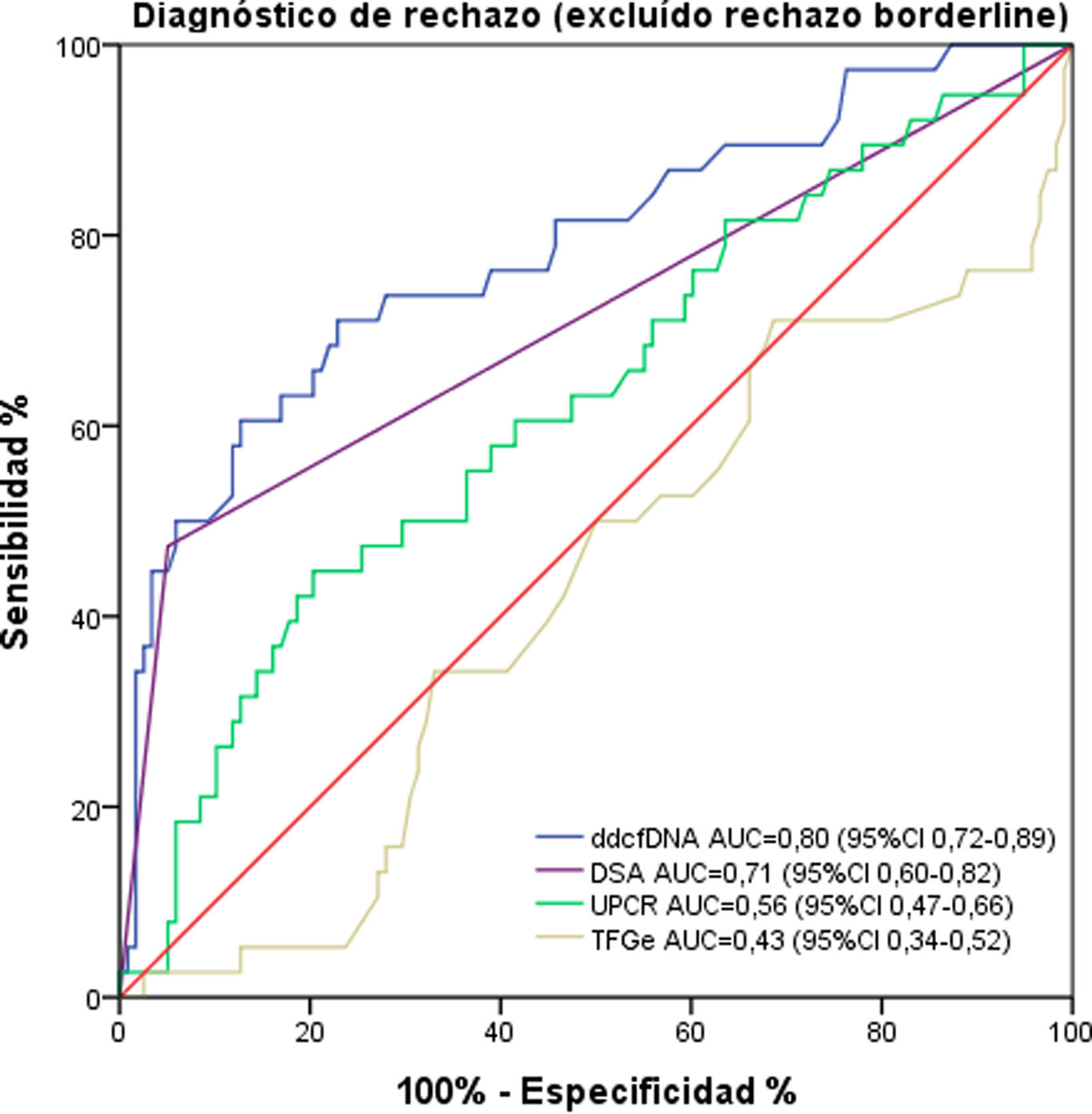
To evaluate the diagnostic performance of ddcfDNA in discriminating between presence and absence of rejection, we calculated the AUC. The result for discriminating any type of rejection (including borderline) from the absence of rejection was 0.74 (0.65–0.82). Excluding borderline rejection from the analysis, the AUC was 0.80 (0.72–0.89) (Fig. 3). For discrimination of ABMR or TCMR exclusively versus no rejection, ddcfDNA exhibited an AUC of 0.84 (0.77–0.92) and 0.59 (0.40–0.77), respectively. If only borderline rejection versus no rejection is compared, an even lower AUC of 0.49 (0.36–0.62) was observed.
ROC curve for ddcfDNA, presence of DSA at biopsy, UPCR and eGFR. ddcfDNA discriminates the diagnosis of rejection (excluding borderline rejection in the analysis) with better performance compared to other measures of renal graft function. ddcfDNA: donor-derived cell-free DNA; DSA: donor-specific antibodies; UPCR: urine protein/creatinine ratio; eGFR: estimated glomerular filtration rate.
The presence of DSA showed an AUC of 0.71 (0.60–0.82), while eGFR and urine protein/creatinine ratio had AUCs of 0.43 (0.34–0.52) and 0.56 (0.47–0.66), respectively (Fig. 3).
DiscussionThe results of this study support the implementation of ddcfDNA as a diagnostic tool in renal posttransplant monitoring. Our data show a statistically significant difference in ddcfDNA levels between patients with and without rejection, with significantly higher levels in those with rejection, especially in antibody-mediated cases. These findings suggest that this biomarker may be more sensitive in the detection of ABMR compared to TCMR or borderline rejection, and are consistent with previous studies,8–10 underscoring the potential of ddcfDNA to identify rejection noninvasively·
In contrast to other studies that have reported higher ddcfDNA levels in severe TCMR rejections (≥IB),11,12 our cohort showed no significant differences between the different TCMR subtypes (P = .788). This could be due to heterogeneity in the presentation and severity of rejection, ddcfDNA not reflecting tissue damage or pathophysiological mechanisms not generating detectable amounts of circulating DNA, in addition to the small sample size of patients with TCMR (n = 11).
This study also supports previous observations on the behavior of ddcfDNA in ABMR,13–15 evidencing significantly higher levels in patients with ABMR than in those with positive DSA but no biopsy damage.
Specifically, patients with ABMR and positive DSA presented a median ddcfDNA of 2.34%, notably higher than that of patients with ABMR without DSA (0.97%; P = .010). The integration of these two types of information (ddcfDNA and presence or absence of DSA) would allow us to discern whether or not the immunological phenomenon is causing graft damage.
In terms of its diagnostic ability, ddcfDNA showed an AUC of 0.74 to discriminate between any type of rejection (including borderline rejection) and the absence of rejection. In particular, its performance was more robust in differentiating ABMR from no rejection, with an AUC of 0.84. These data reflect that ddcfDNA may have limitations in detecting borderline rejection (AUC of 0.49),9,16,17 although in these cases it could help to identify patients with worse prognosis in terms of renal function and rejection progression.11
Comparatively, the diagnostic yield of ddcfDNA was superior to that of other renal function markers such as eGFR and UPCR, which presented AUC of 0.43 and 0.56, respectively. This reaffirms the value of ddcfDNA as a more sensitive and specific marker for acute rejection in renal transplant recipients.
The analysis of ddcfDNA in relation to the individual Banff classification scores supports its utility in assessing ABMR, since it reveals a significant association with microvascular inflammation and CD4 positivity, whereas there was no association with tubulointerstitial inflammation.18–20
Despite these positive results, our study has some limitations. First, the lack of longitudinal follow-up of ddcfDNA levels after biopsy and rejection treatment precludes assessment of its utility as a dynamic prognostic or treatment response biomarker. Previous studies suggest that changes in post-treatment levels could indicate therapeutic response and graft recovery, which this study did not assess.8,16,18 In addition, the heterogeneity of rejection in our sample could have influenced the interpretation of the results. Finally, with the current method for measuring ddcfDNA we cannot differentiate the proportion corresponding to previous non-functioning grafts. However, in our cohort, only one patient presented ddcfDNA levels higher than 1% without rejection data (the patient presented ATN) and the patient was a carrier of a previous graft.20
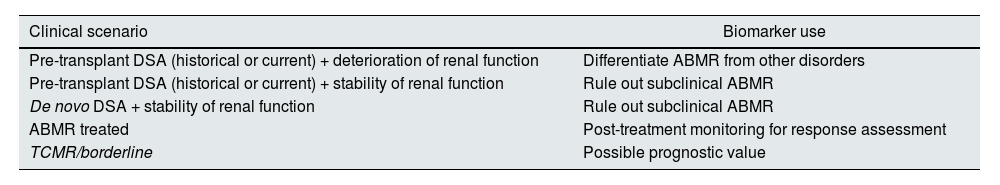
This knowledge allows us to discern where to use the biomarker in different clinical scenarios,11,16,18,21 which we present in Table 4.
Proposal on the use of ddcfDNA according to the different clinical scenario.
| Clinical scenario | Biomarker use |
|---|---|
| Pre-transplant DSA (historical or current) + deterioration of renal function | Differentiate ABMR from other disorders |
| Pre-transplant DSA (historical or current) + stability of renal function | Rule out subclinical ABMR |
| De novo DSA + stability of renal function | Rule out subclinical ABMR |
| ABMR treated | Post-treatment monitoring for response assessment |
| TCMR/borderline | Possible prognostic value |
ABMR: antibody-mediated rejection; ddcfDNA: donor-derived cell-free DNA; DSA: donor-specific antibodies; TCMR: T-cell-mediated rejection.
Finally, the implementation of the test in the hospital itself offers flexibility in obtaining results, adapting to specific clinical needs by allowing the processing of a variable number of samples in each analysis. In addition, direct contact between the nephrologist and the laboratory specialist facilitates discussion and resolution of doubts in complex cases, which favors a deeper understanding of the use of the biomarker. This, in turn, helps to avoid unnecessary biopsies, which not only reduces costs but also optimizes efficiency in clinical management.
In conclusion, this study demonstrates the value of ddcfDNA in the detection of acute rejection in renal transplantation. The significant difference in its levels in ABMR highlights its potential as a diagnostic tool and its advantage over other renal function markers. The hospital implementation of the test facilitates its clinical integration, promoting a timely and accurate diagnosis of rejection, optimizing the management and prognosis of transplanted patients.
FinancingThis study has not received funding. Elena Cuadrado has received an “Emili Letang - Josep Font” research grant from Hospital Clínic de Barcelona.