In this report, we document the CTNS gene mutations of 28 Iranian patients with nephropathic cystinosis age 1–17 years. All presented initially with severe failure to thrive, polyuria, and polydipsia.
MethodsCystinosis was primarily diagnosed by a pediatric nephrologist and then referred to the Iran University of Medical Sciences genetics clinic for consultation and molecular analysis, which involved polymerase chain reaction (PCR) amplification to determine the presence or absence of the 57-kb founder deletion in CTNS, followed by direct sequencing of the coding exons of CTNS.
ResultsThe common 57-kb deletion was not observed in any of the 28 Iranian patients. In 14 of 28 patients (50%), mutations were observed in exons 6 and 7. No mutation was detected in exon 5, and only one (3.6%) patient with cystinosis showed a previously reported 4-bp deletion in exon 3 of CTNS. Four patients (14.3%) had a previously reported mutation (c.969C>A; p.N323K) in exon 11, and five (18%) had novel homozygous deletions in exon 6 leading to premature truncation of the protein. These deletions included c.323delA; p.Q108RfsX10 in three individuals and c.257-258delCT; p.S86FfsX37 in two cases. Other frame-shift mutations were all novel homozygous single base pair deletion/insertions including one in CTNS exon 9 (c.661insT; p.V221CfsX6), and four (14.3%) in exon 4, i.e., c.92insG; p.V31GfsX28 in two and c.120delC; p.T40TfsX10 in two. In total, we identified eight previously reported mutations and eight novel mutations in our patients. The only detected splice site mutation (IVS3-2A>C) was associated with the insertion mutation in the exon 9.
ConclusionThis study, the first molecular genetic analysis of non-ethnic-specific Iranian nephropathic cystinosis patients, may provide guidance for molecular diagnostics of cystinosis in Iran.
En este informe, documentamos las mutaciones del gen CTNS de 28 pacientes iraníes con cistinosis nefropática y una edad de 1-17 años. En un principio, todos presentaron retraso del desarrollo, poliuria y polidipsia.
MétodosEn primer lugar, un nefrólogo pediátrico diagnosticó la cistinosis y luego los pacientes fueron trasladados a la clínica genética de la Universidad de Ciencias Médicas de Irán para consulta y análisis molecular, que incluía la multiplicación por reacción en cadena de la polimerasa (PCR), para determinar la existencia o ausencia de la deleción del fundador del 57kb en el CTNS, seguida por la secuenciación directa de los exones de codificación del CTNS.
ResultadosLa deleción frecuente del 57kb no se observó en ninguno de los 28 pacientes iraníes. En 14 de los 28 pacientes (50%) se observaron mutaciones en los exones 6 y 7. No se detectó ninguna mutación en el exón 5 y solo un paciente (3,6%) con cistinosis mostró una deleción del 4pb, anteriormente comunicada, en el exón 3 del CTNS. De ellos, 4 pacientes (14,3%) tenían una mutación anteriormente comunicada (c.969C > A; p.N323K) en el exón 11 y 5 (18%) tenían nuevas deleciones homocigóticas en el exón 6 que produjeron el vaciamiento prematuro de la proteína. Entre estas deleciones se puede citar c.323delA; p.Q108RfsX10 en 3 personas y c.257-258delCT; p.S86FfsX37 en 2 casos. Otras mutaciones con desplazamiento del marco de lectura fueron todas nuevas deleciones/inserciones de un par de bases únicas homocigóticas, incluyendo una en el exón 9 del CTNS (c.661insT; p.V221CfsX6) y 4 (14,3%) en el exón 4, es decir, c.92insG; p.V31GfsX28 en 2 y c.120delC; p.T40TfsX10 en 2. En total, en nuestros pacientes se identificaron 8 mutaciones anteriormente comunicadas y 8 mutaciones nuevas. La única mutación del sitio de empalme detectada (IVS3-2A>C) estaba asociada con la mutación de inserción en el exón 9.
ConclusiónEste estudio, el primer análisis genético molecular de pacientes iraníes con cistinosis nefropática de carácter no específicamente étnico, puede servir como guía para el diagnóstico molecular de la cistinosis en Irán.
Cystinosis, OMIM# 219800, is a lysosomal storage disease due to impaired transport of cystine across the lysosomal membrane. The gene defect involves CTNS (OMIM 606272; GenBank NM_001031681), located on chromosome 17p13,1 and results in cystine accumulation in most cells and tissues of the body, including the kidneys, eyes, bone marrow, intestines, liver, and spleen. Cystinosis is inherited in an autosomal recessive fashion; all affected individuals have biallelic CTNS mutations.
Cystinosis has been divided into three categories: nephropathic cystinosis, intermediate cystinosis and nonnephropathic or ocular cystinosis, although these subtypes actually reflect a continuum of severity. As the most common and severe phenotype, nephropathic cystinosis manifests in infancy, causing poor growth and renal Fanconi syndrome, characterized by polyuria, polydipsia, proteinuria, acidosis and urinary loss of various salts and nutrients. Renal tubular dysfunction leads to the loss of essential minerals, impairing growth and causing hypophosphatemic rickets. By one year of age, cystine crystals are generally apparent in the corneas on slit lamp examination by an experienced ophthalmologist. These crystals cause pain and increased sensitivity to light.2,3 untreated children experience complete kidney failure by approximately 10 years of age. Long-term complications include muscle deterioration, blindness, inability to swallow, diabetes, hypothyroidism, central nervous system involvement, and male infertility.
The signs of intermediate cystinosis are the same as for nephropathic cystinosis, but they typically appear in adolescence rather than in infancy. Kidney failure and corneal crystals are the most important early features of this disease. If intermediate cystinosis is left untreated, complete kidney failure can occur in the late teens to mid-1920s.3 Individuals with ocular cystinosis typically experience photophobia, but usually do not develop kidney failure or other clinical manifestations of cystinosis. Due to the absence of severe symptoms, the age at which this form of cystinosis is diagnosed varies widely.4,5
Kidney transplantation has significantly extended the lives of patients with nephropathy cystinosis. In addition, the cysteine-lowering drug, cystamine, depletes cells of cysteine by combining with cystine to produce cysteine and cysteine-cystamine mixed disulfide, which exits cystinotic lysosomes via a functional lysine carrier.6 This therapy has been salutary with respect to preservation of renal function, stimulation of growth, and prevention of the late, nonrenal complications of cystinosis.
The CTNS gene has 13 exons, but exons 3–12 are protein-coding; biallelic mutations lead to significant loss of the lysosomal cystine transporter, cystinosin.3,7–9 Approximately 200 CTNS mutations have been reported in cystinosis patients, the most common is a 57-kb deletion responsible for 50% of all the North American and Northern European cases.10–20 Here we report the CTNS mutations present in 28 Iranian patients of diverse ethnicity, providing the first molecular genetic study of cystinosis in this population.
MethodsPatientsStudies were conducted in patients, family members and normal control subjects after informed consent was obtained according to Iran University ethical committee codes. Blood samples were collected from individuals with nephropathic cystinosis and their parents who were attending Ali-Asghar Children Hospital of Iran University of Medical Science. The clinical and molecular genetics survey was performed in accordance with the Declaration of Helsinki.
During two years of study, 28 individuals (16 males and 12 females) from 28 unrelated consanguineous families participated in our studies. The patients were from different Persian ethnic groups of Iran, and inclusion required signs and symptoms of Fanconi syndrome and/or the presence of cystine crystals in the cornea, documented by a nephrologist and/or ophthalmologist, respectively. All patients had a clinical history consistent with the disease, including kidney and/or eye involvement; patients were generally referred to our clinic for genetic counseling and molecular genetic analysis by pediatric nephrologists.
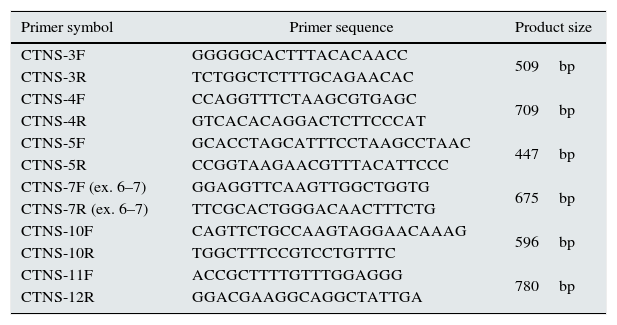
PCR amplification of CTNSGenomic DNA was extracted as previously described.7 The 5′ UTR and the open reading frame (ORF) of CTNS (with the exon–intron boundaries) were amplified by independent PCR amplifications utilizing exon-specific primer pairs, which the sequences of some are listed in Table 1. PCR was performed in a final volume of 50μl containing 200μM dNTP, 1×Taq DNA polymerase buffer, various concentration of MgCl2, 2U Taq proof-reading (Pfu) DNA polymerase (Cinagen, Tehran, Iran), 200ng genomic DNA and 10pmol of each primer. After initial denaturation for 5min at 95°C, 35 cycles of amplification were performed as follows: 30s at 95°C, 30s at 51–57°C (depends on the primer pairs) and 30s at 72°C. PCR products were evaluated on a 1.5% agarose gel stained with RedSafe (Cinagen, Tehran, Iran).
CTNS gene-specific primers.
| Primer symbol | Primer sequence | Product size |
|---|---|---|
| CTNS-3F | GGGGGCACTTTACACAACC | 509bp |
| CTNS-3R | TCTGGCTCTTTGCAGAACAC | |
| CTNS-4F | CCAGGTTTCTAAGCGTGAGC | 709bp |
| CTNS-4R | GTCACACAGGACTCTTCCCAT | |
| CTNS-5F | GCACCTAGCATTTCCTAAGCCTAAC | 447bp |
| CTNS-5R | CCGGTAAGAACGTTTACATTCCC | |
| CTNS-7F (ex. 6–7) | GGAGGTTCAAGTTGGCTGGTG | 675bp |
| CTNS-7R (ex. 6–7) | TTCGCACTGGGACAACTTTCTG | |
| CTNS-10F | CAGTTCTGCCAAGTAGGAACAAAG | 596bp |
| CTNS-10R | TGGCTTTCCGTCCTGTTTC | |
| CTNS-11F | ACCGCTTTTGTTTGGAGGG | 780bp |
| CTNS-12R | GGACGAAGGCAGGCTATTGA |
Sequence analysis of PCR products from the promoter region and all exons was performed after purification of PCR products (PCR product purification kit, Roche). Both strands were sequenced using the Big Dye Termination system in an ABI 310 capillary sequencer (Macrogen, South Korea). Sequencing results were analyzed using bioinformatic tools (http://www.ncbi.nlm.nih.gov/gorf/bl2.html).
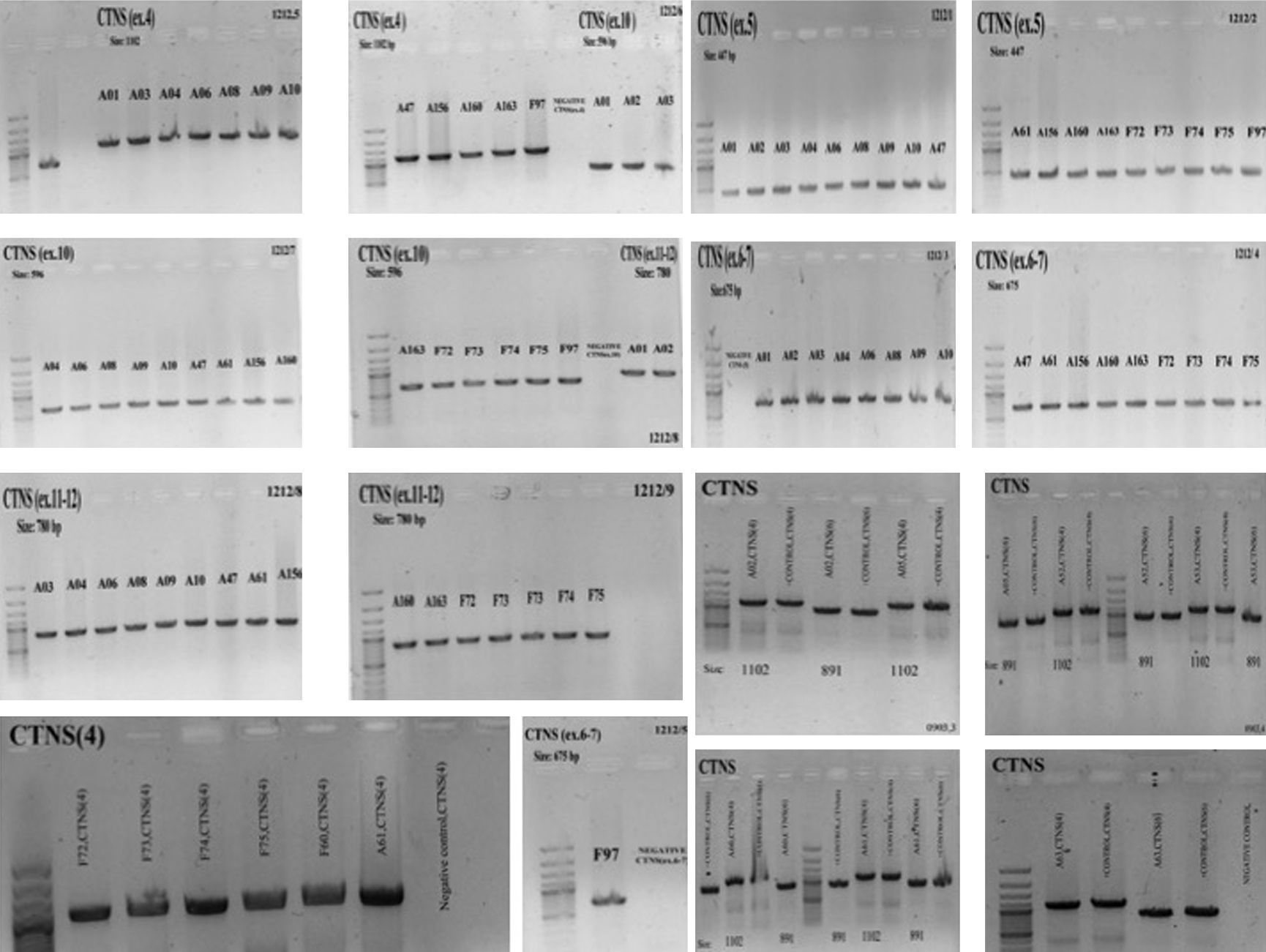
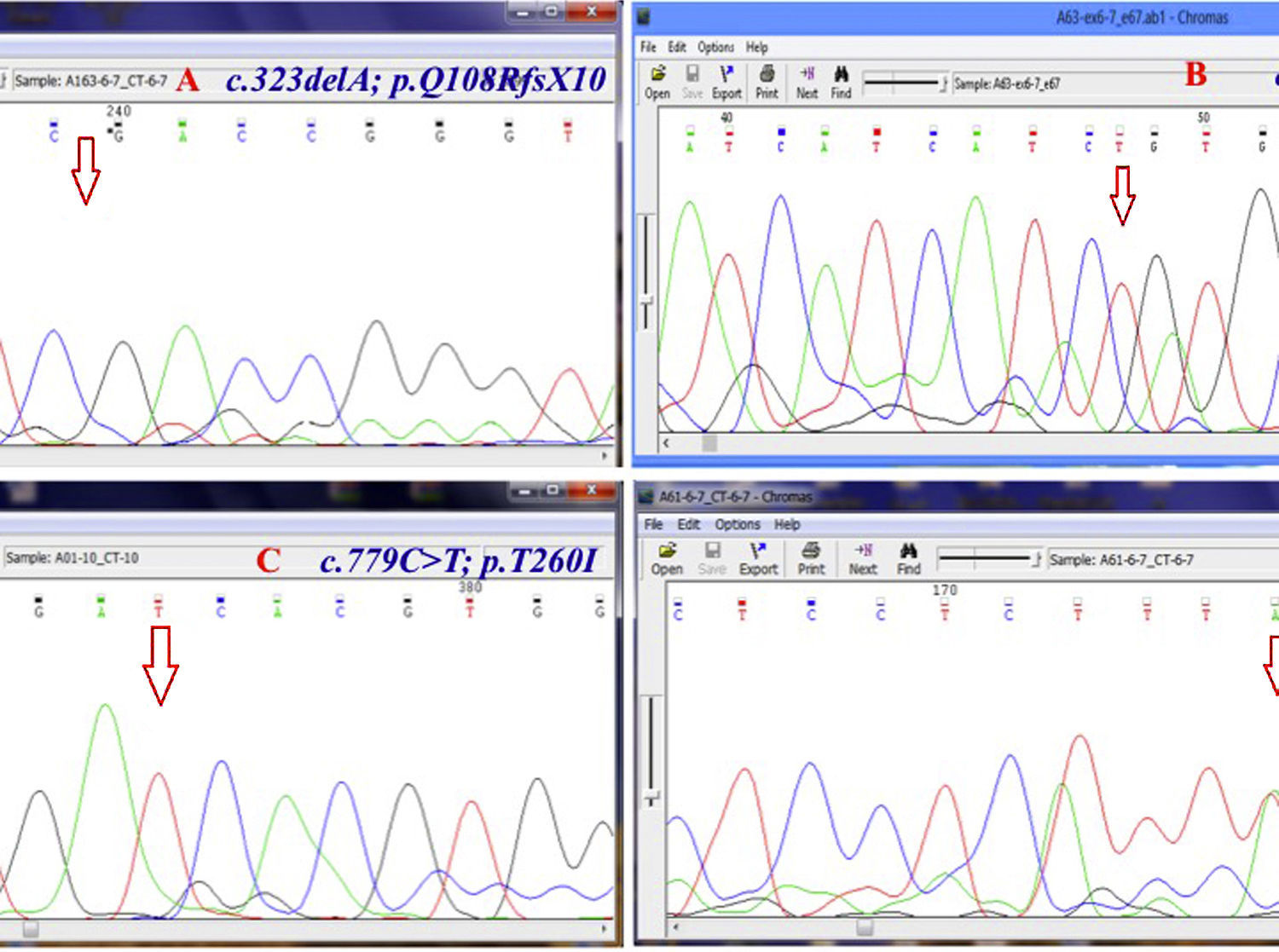
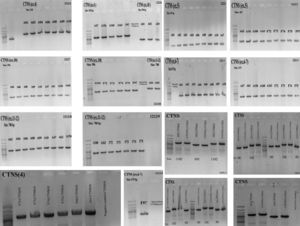
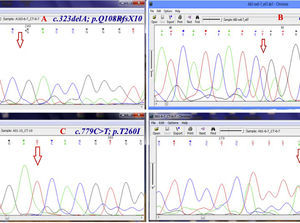
ResultsGAP-PCR was carried out in all samples, and no patient showed the 57kb CTNS deletion in either the homozygous or heterozygous state (Fig. 1). Direct sequencing of the entire CTNS gene demonstrated alterations in all patients. In 14 of 28 patients (50%), the alterations were observed in exons 6 and 7. Four patients (14.15%) had c.969C>A; p.N323K, in exon 11 of the CTNS gene which was previously reported, 3 were heterozygous and one was homozygous. Three patients (11%) were homozygous for a single base pair deletion in nucleic acid position 323 (c.323delA) in exon 6, leading to premature truncation of the protein (p.Q108RfsX10) (Fig. 2A). Also in exon 6, two patients (7.2%) showed homozygosity for an unreported deletion of two nucleotides, (c.257-258delCT; p.S86FfsX37). Two patients were homozygous for a novel single base pair insertion (c.92insG; p.V31GfsX28) in exon 4, and two others had a homozygous deletion (c.120delC; p.T40TfsX10) in exon 4. Another frame-shift mutation was identified as a novel homozygous single base pair deletion in exon 9 (c.661insT; p.V221CfsX6) in one patient (Fig. 2B). Also, one patient had a previously reported mutation (http://www.ncbi.nlm.nih.gov/clinvar/variation/188834/), c.18-21del GACT; p.T7fsX7, in CTNS exon 3. The variations in -303 5′ UTR of the CTNS, previously reported for ocular cystinosis,15 were detected in 2 patients in our study too.
Genomic PCR amplifies CTNS amplicons. Genomic PCR amplifies the exons using the gene-specific pairs of primers. The common 57-kb deletion was not observed in any of the 28 Iranian patients. The PCR products were subjects for Sanger sequencing using relevant forward and reverse primers after purification of PCR products (PCR product purification kit, Roche).
Chromatogram (Chromas software version 2.4.1) for the region containing some novel mutations. Red arrow shows the substituted nucleotide. Sequence analysis of PCR products from the promoter region and all exons was performed. Both strands were sequenced using the Big Dye Termination system in an ABI 310 capillary sequencer (Macrogen, South Korea). A, shows a single base pair deletion in nucleic acid position 323 (c.323delA) in exon 6, leading to premature truncation of the protein p.Q108RfsX10 which was observed in three patients (11%) in homozygous state. B, c.661insT; p.V221CfsX6 is a novel homozygous single base pair deletion in exon 9 observed in one patient. C, the variant c.779C>T; p.T260I (in exon 10) was detected in 18 of 28 (64%) patients. D, c.261T>A; p.F87L in exon 6 of CTNS is the most common mutation in our population which was observed in six patients (21.4%) in heterozygote and homozygote states.
In our population, the variant c.779C>T; p.T260I (in exon 10) was detected in 18 of 28 (64%) patients with nephropathic cystinosis (Fig. 2C). Furthermore, a second CTNS mutation was detected in 16 of 18 patients. The most common mutation in our population was c.261T>A; p.F87L in exon 6 of CTNS; this was observed in six patients (21.4%) (Fig. 2D).
Five patients were compound heterozygous for the mutations c.73A>T; p.S25C/c.261T>A; pF87L, c.969C>A; p.N323K/c.1015G>A; p.G339R, c.73A>T; p.S25C/c.969C>A; p.N323K, and c.261T>A; pF87L/c.969C>A; p.N323K.
A summary of all identified mutations is depicted in Table 2. In addition, a number of SNPs were found in coding exons of the CTNS gene (Table 3). A total of 68 healthy non-related Persian individuals were screened for the novel mutations found in this survey; none carried any of the changes.
Summarized mutations detected in patients with cystinosis in Iran. All variations have been verified with the mutations reported in ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/?term=ctns%5Bgene%5D).
| No. | Amplicone (size) Patient code | Exon 1–2 (809bp) | Exon 3 (509bp) | Exon 4 (121bp) | Exon 5 (85bp) | Exon 6–7 (1373bp) | Exon 8–9 (291bp) | Exon 10 (–bp) | Exon 11–12 (118bp) | Description |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A01 | c.119C>G; p.T40S (Het) c.124G>A; p.V42I (Het) | c.779C>T; p.T260I | c.969C>A; p.N323K (Het) | Juvenile nephropathic cystinosis | |||||
| 2 | A02 | c.250A>G; p.N84D (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 3 | A03 | c.73A>T; p.S25C | c.261T>A; p.F87L | c.779C>T; p.T260I | Novel (Comp. Het) | |||||
| 4 | A04 | c.73A>T; p.S25C | c.261T>A; p.F87L | c.779C>T; p.T260I | Novel (Comp. Het) | |||||
| 5 | A05 | c.92insG; p.V31GfsX28 (Hom) | Novel | |||||||
| 6 | A06 | c.779C>T; p.T260I (Hom) | IVS11+70T | |||||||
| 7 | A08 | c.194A>G; p.K65R (SNP) | c.779C>T; p.T260I | c.969C>A; p.N323K (Het) c.1015G>A; p.G339R (Het) | Juvenile nephropathic cystinosis (Comp. Het) | |||||
| 8 | A09 | -303 TG Ins 5′ UTR (Hom) | c.779C>T; p.T260I | Ocular cystinosis | ||||||
| 9 | A10 | c.120delC; p.T40TfsX10 (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 10 | 19 | c.647C>G; p.T216R19 (Hom) | Novel | |||||||
| 11 | A47 | c.261T>A; p.F87L (Hom) | Novel | |||||||
| 12 | A51 | -303 G>T 5′ UTR | Ocular cystinosis | |||||||
| 13 | A52 | c.120delC; p.T40TfsX10 (Hom) | Novel | |||||||
| 14 | A53 | c.92inserG; p.V31GfsX28 (Hom) | Novel | |||||||
| 15 | A60 | c.18-21del GACT; p.T7fsX7 (Hom) | – | |||||||
| 16 | A61 | c.261T>A; p.F87L (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 17 | A63 | IVS3-2A>C | c.661insT; p.V221CfsX6 (Hom) | Novel | ||||||
| 18 | A156 | c.250A>G; p.N84D (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 19 | A160 | c.73A>T; p.S25C | c.779C>T; p.T260I | c.969C>A; p.N323K | Juvenile nephropathic cystinosis (Comp. Het) | |||||
| 20 | A163 | c.323delA; p.Q108RfsX10 (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 21 | F72 | c.323delA; p.Q108RfsX10 (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 22 | F73 | c.261T>A; p.F87L (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 23 | F74 | c.257-258delCT; p.S86FfsX37 (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 24 | F75 | c.261T>A; p.F87L | c.779C>T; p.T260I | c.969C>A; p.N323K (Het) | Juvenile nephropathic cystinosis (Comp. Het) | |||||
| 25 | F80 | c.257-258delCT; p.S86FfsX37 (Hom) | Novel | |||||||
| 26 | F97 | c.779C>T; p.T260I (Hom) | ||||||||
| 27 | F129 | c.261T>A; p.F87L (Hom) | c.779C>T; p.T260I | Novel | ||||||
| 28 | F616 | c.323delA; p.Q108RfsX10 (Hom) | Novel |
Summary of CTNS mutations in cystinosis (by ClinVar) (http://www.ncbi.nlm.nih.gov/clinvar/?term=ctnsall).
| Variation Location | Condition(s) | Clinical significance (last reviewed) |
|---|---|---|
| CTNS, 1-BP INS, -303T | Cystinosis, ocular nonnephropathic | Pathogenic (October 1, 2001) |
| CTNS, -303G-T | Cystinosis, ocular nonnephropathic | Pathogenic (October 1, 2001) |
| CTNS, -295G-C | Cystinosis | Pathogenic (October 1, 2001) |
| CTNS, 5-BP DEL, NT545 | Cystinosis | Pathogenic (November 1, 1998) |
| CTNS, IVS7AS, C-G, -10 | Juvenile nephropathic cystinosis | Pathogenic (December 1, 1999) |
| CTNS, 4-BP DEL, 18GACT | Juvenile nephropathic cystinosis, Cystinosis | Pathogenic (November 1, 2009) |
| CTNS, 2-BP DEL, 397TG | Cystinosis | Pathogenic (April 1, 1998) |
| NC_000017.11:g.3600934_3658165del57232 | Cystinosis, ocular nonnephropathic, Juvenile nephropathic cystinosis | Pathogenic (August 11, 2011) |
| NM_080704.3(TRPV1):c.-34+7395T>C | Cystinosis | Benign (January 14, 2015) |
| NM_004937.2(CTNS):c.18_21delGACT (p.Thr7Phefs) | Cystinosis | Pathogenic (April 24, 2015) |
| NM_001031681.2(CTNS):c.124G>A (p.Val42Ile) | Cystinosis, atypical nephropathic | Pathogenic (December 1, 1999) |
| NM_001031681.2(CTNS):c.198_218del21 (p.Ile67_Pro73del) | Cystinosis | Pathogenic (July 31, 2014) |
| NM_001031681.2(CTNS):c.199_219del21 (p.Ile67_Pro73del) | Cystinosis | Likely pathogenic (July 31, 2014) |
| NM_004937.2(CTNS):c.225+5_225+6delGTinsCC | Cystinosis | Likely pathogenic (July 21, 2014) |
| NM_001031681.2(CTNS):c.283G>T (p.Gly95Ter) | Cystinosis | Pathogenic (April 1, 1998) |
| NM_004937.2(CTNS):c.292dupA (p.Thr98Asnfs) | Cystinosis | Likely pathogenic (April 3, 2014) |
| NM_001031681.2(CTNS):c.329G>T (p.Gly110Val) | Cystinosis, atypical nephropathic | Pathogenic (December 1, 2002) |
| NM_001031681.2(CTNS):c.356G>A (p.Arg119His) | Not specified | Benign (April 15, 2015) |
| NM_004937.2(CTNS):c.414G>A (p.Trp138Ter) | Cystinosis | Pathogenic (April 24, 2015) |
| NM_004937.2(CTNS):c.416C>T (p.Ser139Phe) | Juvenile nephropathic cystinosis, atypical nephropathic, cystinosis | Pathogenic (May 30, 2014) |
| NM_004937.2(CTNS):c.473T>C (p.Leu158Pro) | Cystinosis | Pathogenic (August 11, 2011) |
| NM_001031681.2(CTNS):c.506G>A (p.Gly169Asp) | Cystinosis | Pathogenic (November 1, 1998) |
| NM_004937.2(CTNS):c.559_561+24del27 | Cystinosis | Pathogenic (February 15, 2015) |
| NM_004937.2(CTNS):c.561+1delG | Cystinosis | Likely pathogenic (November 24, 2014) |
| NM_004937.2(CTNS):c.589G>A (p.Gly197Arg) | Cystinosis, ocular nonnephropathic, cystinosis | Pathogenic (March 3, 2014) |
| NM_001031681.2(CTNS):c.612C>T (p.Asn204=) | Not provided | Uncertain significance (March 30, 2015) |
| NM_004937.2(CTNS):c.613G>A (p.Asp205Asn) | Cystinosis | Pathogenic (August 11, 2011) |
| NM_004937.2(CTNS):c.614_616delACG (p.Asp205del) | Cystinosis | Likely pathogenic (March 12, 2014) |
| NM_004937.2(CTNS):c.696dupC (p.Val233Argfs) | Cystinosis | Pathogenic (August 11, 2011) |
| NM_004937.2(CTNS):c.809_811delCCT (p.Ser270del) | Cystinosis | Likely pathogenic (October 16, 2014) |
| NM_004937.2(CTNS):c.853-3C>G | Cystinosis, ocular nonnephropathic | Pathogenic (August 11, 2011) |
| NM_004937.2(CTNS):c.926dupG (p.Ser310Glnfs) | Cystinosis | Likely pathogenic (March 7, 2014) |
| NM_001031681.2(CTNS):c.969C>G (p.Asn323Lys) | Juvenile nephropathic cystinosis | Pathogenic (August 1, 1999) |
| NM_001031681.2(CTNS):c.1015G>A (p.Gly339Arg) | Cystinosis | Pathogenic (September 1, 2001) |
The incidence of cystinosis is estimated to be between 1 in 100,000 and 1 in 200,000 worldwide; it is predicted to be much higher in Iran due to the nation's high rate of consanguinity. The CTNS gene spans 26.6kbp and contains 13 exons. A 57,257-bp deletion8 was found to be responsible for approximately half of Northern European and North American cystinosis patients10,11; this deletion is easily detected by a multiplex PCR amplification assay.21 Cystinosis has been reported in different ethnic groups, and about 200 different mutations have been described to date, including mutations in the promoter region, missense, nonsense, deletion, insertion, and splice-site mutations.9–11,22 However mutational spectrum of CTNS in African, Mediterranean, and middle east populations seems to be different (Table 4). Meanwhile in a population of Saudi Arabian origin, it showed small deletions and point mutations.24 None of the Egyptian patients with infantile nephropathic cystinosis who underwent molecular analysis had the 57-kb deletion,24 consistent with a study in Turkey.12 These patients showed frameshift, nonsense, missense, and intronic splicing mutations. Previous studies of nephropathic cystinosis in patients of unrelated families from southwest Iran also showed point mutations.13 Also we have not found this CTNS large deletion in any of screened Iranian patients with cystinosis. Since the 57-kb deletion originated approximately 1500 years ago in northern Europe,9 it is not surprising that it is absent among Iranian cystinosis patients.
Summarized previous reports of genetic diagnosis of cystinosis in African and Mediterranean countries.
| References | ||
|---|---|---|
| African–American origin | ||
| (IVS 5+5 GT→CC) | Homozygous | 17 |
| 57-kb deletion | Homozygous | |
| 928G→A 57-kb deletion | Heterozygous | |
| Tunisian origin | ||
| c.771_793del (p.Gly258Serfs*30) | 15 | |
| 20-kb deletion, Deleted intron 1 CARKL–intron 6 CTNS | ||
| c.1515G>A (p.G308R) | Homozygous | |
| c.771_793del (p.Gly258Serfs*30) | Homozygous | |
| Spanish origin | ||
| 416C-T transition (S139F) | Compound heterozygous | 19 |
| GACT del, at nucleotide 357 | ||
| Italian origin | ||
| 569–577delTTCTCCTCA | 7 | |
| c.922G>A | ||
| c.681+1G>A | ||
| c.696–697insC | ||
| c.861G>A | ||
| c.1015G>A | ||
| c.225+3A>T | ||
| c.890G>A | ||
| c.659–665delTCGTGCA | ||
| c.18-21delGACT | ||
| c.699-700delGT | ||
| 1366del12bp | ||
| -295G>C | ||
| Egyption origin | ||
| Exons 6, c.260_261delTT (p.F87SfsX36) Exon 8, c.560A>G (p.K187R) | Heterozygous | 23 |
| Exons 10, c.734G>A (p.W245fsX) Exon 12, c.1032delCinsTG (p.F345CfsX19) | Heterozygous | |
| Exon 12, c.1084G>A; pG362R | Homozygous | |
| IVS3+5g>t | ||
| 1-593-41C>T | ||
| Exon 10,c.829dup (p.T277NfsX19) | Homozygous | |
| Exon 11, c.922G>A (p. G308R) | Homozygous | |
| Exon 10, 809_811del (p.S270del) | Homozygous | |
| Exon 3, c.15G>A (p.W5X) | Homozygous | |
| Exon 9, c.681G>A (E227E) | Homozygous | |
| Exon 12, c.1015G>A | Homozygous | |
| Turkish origin | ||
| c.1015 G>A; p.G339R c.451A>G; p.R151G | Heterozygous | 12 |
| c.18_21delGACT; p.T7FX7 | Homozygous | |
| 18_21delGACT;p.T7X7 c.470 G>A; p.G157D | Heterozygous | |
| c.518A>G, p.Y173C | Homozygous | |
| c.470 G>A; p.G157D | Homozygous | |
| c.140+1 G>T | Homozygous | |
| Moraco | ||
| c.61+5G>A (5′ splice site IVS3) | 19 | |
| c.922G>A (p.G308R) | ||
| Jordanian origin | ||
| c.779C>T (p.Thr26Ile) | 8 | |
| exon 10, c.829dupA (p.Thr277Asnfs*19) | ||
| Exon 11, c.890G>A (p.Trp297X). | ||
| Iranian origin | ||
| Exon 1–2 -303TG Ins 5′UTR -303 G>T 5′UTR | This Study | |
| Exon 3 c.18-21del GACT (p.T7fsX7) | ||
| Exon 4 c.119C>G (p.T40S) c.124G>A (p.V42I) c.73A>T (p.S25C) c.92insG (p.V31GfsX28) c.120delC (p.T40TfsX10) IVS3-2A>C | Heterozygous | |
| Exon 5 c.194 A>G (p. K65R) | Heterozygous | |
| Exon 6–7 c.250A>G (p.N84D) c.261T>A (p. F87L) c.323 del A (p. Q108RfsX10) c.261T>A (p. F87L) c.257–258 del CT (p.S86F fsX37) | Heterozygous | |
| Exon 8–9 c.647C>G (p.T216R) c.661insT (p.V221CfsX6) | Heterozygous | |
| Exon 10, c.779C>T (p.260I) | Heterozygous | |
| Exon 11–12 c.969C>A (p.N323K) IVS 11+70T c.1015G>A (p.G339R) | Heterozygous | |
In this study, most of the patients showed novel mutations although some previously reported mutations have also been detected. Moreover, half of the mutations have been observed in the exons 6 and 7 of the CTNS gene, which is highly suggestive of genetic analysis of these two exons by Sanger sequencing as Tier 1 in Iranian population. Utilizing this recommendation in suspected patients with nephropathic cystinosis in Iran, the process of mutation detection is facilitated, and the cost and time of CTNS mutation detection may be reduced.
The c.779C>T was primarily described in a heterozygous genotype in 5 Italian and some other Jordanian patients.7,8 Moreover, its association with some mutant alleles has been introduced as an additional DNA marker for prenatal molecular diagnostics of cystinosis in Jordanian families.8 Also G339R, one detected mutation in a patient, has already been proposed as a common cause of nephropathic cystinosis in the south western Ontario Amish Mennonite population.20 Overall, all 28 patients in this study including the ones with novel mutations presented with Fanconi syndrome between 6 and 24 months old, and also developed eye involvement. So we admit that clinical feature of the studied patients was consistent with and similar to what is already mentioned in the literature.
Conflict of interestThe authors declare no competing financial interest.
This study was approved and supported by grant no. 93-04-30-25293 by Research Deputy, IUMS. It was also supported by the Intramural Research Program of NHGRI. Authors are thankful of Mrs Farzaneh Vali for her technical support.